Evidence from microbial abundance surveys across the globe suggest that microbes often (or even primarily) live in surface-associated communities, which are typically referred to as biofilms, or spatially structured microbial communities. Within these communities, metabolic activity of the cells creates gradients of nutrients and waste products, which leads to micron-scale physiological heterogeneity that emerges as a direct consequence of microbial growth. Understanding the development and consequences of this heterogeneity in multi-species communities is a major current challenge in microbiology, with important applications for controlling the microbiota, e.g., in our mouth and our digestive tract.
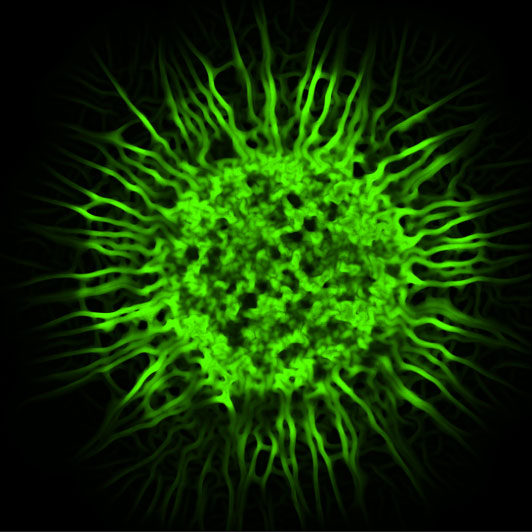
Figure: Confocal microscopy image of a biofilm colony of Escherichia coli cells. The BiofilmQ software enables quantitative phenotyping of such biofilms, and the characterization of properties inside biofilms with spatial and temporal resolution.
Due to the spatial organization of cellular phenotypes and genotypes in microbial communities, studies often rely on microscopy to investigate these communities. Therefore, quantitative image analysis tools for microbial communities are essential. However, modern tools for the comprehensive analysis of spatial heterogeneities in microscopy images of microbial communities were lacking.
To address this problem, we had developed numerous individual image analysis scripts over the years, to provide fixes for our own image analysis needs in previous projects. We have now merged all of these existing image analysis tools, expanded them dramatically, and generalized them to be able to offer image analysis for 2D and 3D communities with time series, and we developed a user-friendly interface. The resulting software, BiofilmQ, now offers a comprehensive analysis workflow, from the import of the raw data to the generation of a multitude of different graphs and visualizations of the data.
The HFSP Career Development Award (CDA) funding to Knut Drescher was instrumental in getting this project started. Often, it is difficult to get method-development projects funded, even if these methods are highly sought-after in the scientific community. This tool emerged as a side-result of the CDA project.


































