Although antibiotics interfere with the functioning of well-characterized molecular targets inside bacteria, we still know very little of the mechanisms that ultimately lead to cell death. Mounting evidence suggests that the action of antibiotics on their targets can promote system-level perturbations, for example, by altering gene expression, cell physiological state and composition, which in turn can ‘push’ vital functions past their tipping points. Molecular targets can be very different, ranging from DNA, to protein biosynthesis, to cell wall, etc. but the global physiological response of a cell in presence of these perturbations can be similar. In this work, we explored the effects that sublethal concentrations of antibiotics have on the dynamics of genome and cytoplasm. We found that both genome and cytoplasm dynamics are altered by the drugs in similar ways, and these changes are consistent with how antibiotics affect the levels of molecular crowding, suggesting a causal link between the two phenomena.
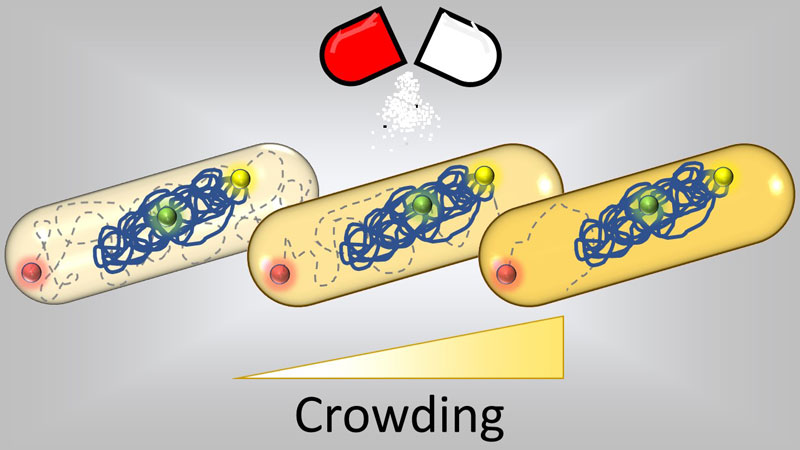
Figure: under antibiotic treatment, both cytoplasmic and chromosomal motion react coherently to the changes in molecular crowding caused by the drug.
To draw a picture of the dynamics during antibiotic treatment that would capture the whole spectrum of motion, we fluorescently tagged two distant loci on the genome, one near the origin and one near the terminus of replication. After having supplemented cells with antibiotics, we followed these fluorescent foci for 10-second intervals over two hours. Tracking of foci position over time revealed that antibiotics that target different aspects of bacterial physiology produce different genomic dynamics. Of the antibiotics tested, ciprofloxacin produced the most marked effect, with loci moving faster than in untreated cells. As a further comparison, we treated a subset of samples with sorbitol, a sugar that E. coli cannot metabolize and that induces a hyperosmotic shock, thereby reducing cell volume. We speculated that this perturbation would increase intracellular density and thus, potentially, reduce intracellular motility. Indeed, cells treated with sorbitol showed significantly slower dynamics. The dynamics of the E. coli chromosome are known to vary depending on the position of a locus with respect to the origin of replication, with closer loci showing larger motions. In all our observations, differences in motility were consistent between the origin and the terminus, suggesting that a common cause affected the motion of distant loci.
We then wondered whether the observed changes in loci dynamics were also witnessed by the cytoplasm, as this would suggest the involvement of a general mechanism at the cell level. To test cytoplasmic dynamics, we made the cells produce fluorescent cytoplasmic particles, made from viral capsids tagged with fluorescent proteins, and we tracked their motility as we had done with the chromosomal loci. Once again, the results for cytoplasmic particles recapitulated our observations on the chromosome, indicating a global correlation between genome and cytoplasmic motility.
Next, we sought to investigate the mechanism that was inducing these changes at the cell level. We started by testing whether our hypothesis that the slower intracellular dynamics of cells treated by sorbitol were driven by increased molecular crowding. By using optical density measurements, refractive index and dry mass estimations we confirmed that this was the case, as sorbitol-treated cells had a significantly higher density than untreated ones. We thus measured intracellular density as a proxy for crowding level also in antibiotic treated cells. Our findings are consistent with the hypothesis that crowding drives both the dynamics of cytosol and chromosome in antibiotic treated bacteria, therefore unveiling a previously unknown system-level effect that is shared by drugs of different classes.
The results of this study, which includes theoretical, advanced data-analysis, and experimental components that could come together thanks to our HFSP collaboration, are fundamental, but also have important implications for treatment design and the development of novel therapeutics. For example, as it emerges from our study, osmotically active substances could act on the same system-level aspects of certain antibiotics, opening the door to potential synergistic or antagonistic effects.


































