Fungi from the genus Candida are common, harmless inhabitants of our bodies. However, when we become immunocompromised, they can cause a variety of diseases, from superficial infections to life threatening systemic conditions. The ability of these microorganisms to adhere to surfaces and form communities known as biofilms allows them to colonize a variety of niches in the human host. In the most medically relevant species, Candida albicans, the formation of biofilms is regulated by a complex network composed by seven regulatory genes that control over 1,000 target genes. Despite its complexity, several findings suggested that this gene regulatory network has changed rapidly in recent evolutionary time. To understand how this network came about, we analyzed biofilm formation in many fungal species and characterized the gene networks responsible for this trait in four Candida species that last shared a common ancestor about 70 million years ago.
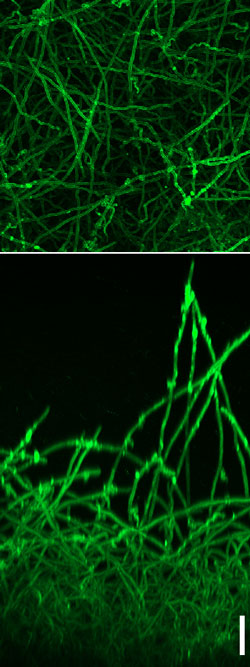
Figure: C. albicans biofilm as observed under the confocal microscope. The biofilm is formed by a basal layer of cells with yeast morphology and a thick upper layer of filamentous cells. The lower panel shows the side-view and the upper panel, the top-view of the biofilm. The scale bar represents 50 μm.
We found that only close relatives can form biofilms similar to C. albicans. Furthermore, even in the species that form biofilms similar in thickness and structure, the gene regulatory networks that control this process have changed. While the DNA binding affinities of regulatory genes have remained conserved through time, their target genes have evolved rapidly: most of the changes in the networks were due to modifications in the cis regulatory regions adjacent to the target genes. Despite the observed differences, the networks in the four species have “structural” similarities. For example, the target genes in all the four networks are highly connected to the regulatory genes, and most regulatory genes seem to regulate themselves and each other. Our work sheds light on the evolution of Candida biofilm formation and how biofilms are orchestrated in these important human pathogens.
This research was made possible by an HFSP Long-Term Fellowship, which enabled collaboration between experts in yeast functional genomics and Candida biology and evolution.


































