 |
Kotaro Fujii obtained his PhD in RNA biochemistry, when he elucidated the molecular mechanism that maintains functional protein production by eliminating aberrant ribosomes. As he enjoys a challenge, he then decided to change country and research field and apply for an HFSP postdoctoral fellowship in the lab of Maria Barna at Stanford University. These changes enabled him to employ his expertise in the context of developmental biology and evolution and led him to discovering dynamic translational regulation in developing mouse embryos and dynamic translation fidelity across evolution. He is naturally curious and aims to reveal the novel molecular mechanism of gene regulation during mRNA translation. |
The ribosome is an ancient molecular machine responsible for translating the genetic code, which has a core ribosomal RNA (rRNA) secondary structure that is universally shared across all kingdoms of life. However, due to 30 insertions of eukaryote specific expansion segments (ESs) in the rRNA, there have been large expansions in the size of the ribosome across evolution. ESs are enigmatic insertions, the longest of which resemble tentacle-like extensions that vary in length and sequence across evolution. Whether ESs have a functional role in translational control and the significance of their dramatic expansions in evolution remain largely unknown.

Cover story Molecular Cell, 728, 20 December 2018.
By selectively engineering rRNA in yeast, we find that one of the largest ESs, ES27L, has an unexpected function in translation fidelity. Ribosomes harboring a deletion in the distal portion of ES27L have sensitivity to the translation error-inducing drug, Paromomycin, that is mutant decreased translation fidelity by increasing amino acid misincorporation, as well as read-through and frameshift errors. Low translation fidelity and accumulation of translation errors can reduce the levels of functional proteins and foster protein aggregation, and may underlie neurological disease. Although complex organisms have higher translational fidelity to produce longer proteins and more complex proteome, how the ribosome itself has increased the precision of protein synthesis across evolution remains an outstanding question.
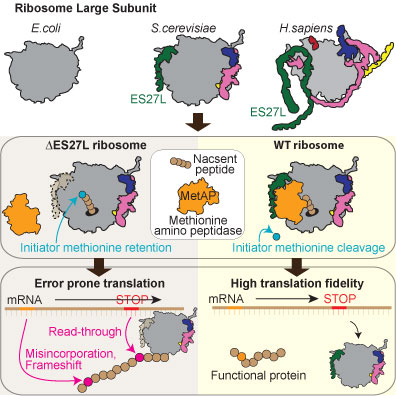
Figure: Graphic model showing the change in the ribosome large subunit across species with a different color for each expansion segment (ES). Selective engineering of the ES27L in yeast revealed that ES27L worked as RNA scaffold to recruit conserved enzyme Methionine amino peptidase (MetAPs) close to the nascent peptide exit tunnel to process initiator methionine (iMer) cleavage, which has a critical role to increase translation fidelity by reducing amino acid misincorporation, stop codon read-through, and frameshift.
By employing quantitative mass spectrometry between WT and ES27L mutants, we further found that ES27L acts as a RNA scaffold to facilitate binding of a conserved enzyme, methionine amino peptidase (MetAP). In all kingdoms of life, translation starts from initiator methionine (iMet), and iMet is subsequently cleaved off co-translationally from almost one-third of all proteins in eukaryotes and prokaryotes by MetAPs. We show that MetAP unexpectedly controls the accuracy of ribosome decoding, which is coupled to an increase in its enzymatic function through its interaction with ES27L. Interestingly, the strong association between MetAP and the ribosome was not observed in the Prokaryote, which does not possess ES27L. These findings suggest that ES27L may be holding the MetAP family of proteins close to the exit tunnel specifically in the eukaryotic ribosome, consistent with the N-terminal processing functions of MetAPs and increasing translation fidelity.
These findings reveal that variable ESs of the ribosome could serve important functional roles and act as platforms for the binding of factors that modulate translation across evolution. In complex organisms, accurate gene expression in time and space is the key to proper cell differentiation and body patterning. Indeed, my previous work revealed the extensive translational regulation of developmental signaling circuitries in developing mouse embryos (Fujii et al., 2017). To achieve such complex regulation in time and space, the enigmatic changes of ESs may endow this ancient molecular machinery, the ribosome, with greater regulation.


































