Cells maintain sophisticated gene expression patterns that regulate all cellular functions. These complex genetic networks must be evolutionarily flexible if cells are to cope with changes in their environment. Indeed, in the course of evolution, the genetic elements that regulate the timing and magnitude of gene expression change according to cellular needs. These regulatory elements are called promoters, they are coded in the DNA right next to genes, and they determine which genes are expressed by recruiting the transcription machinery to the genes under their control.
How new promoters evolve is a central question that is not completely understood. For example, a typical promoter in the bacterium E. coli contains two sequence features, six bases each, that are required for the transcription machinery in order to bind and transcribe genes. One might think that it would be difficult for E. coli to evolve a new promoter from scratch. In fact, exactly the opposite is true. In a recent study Avihu Yona and colleagues revealed a new aspect of promoter evolution by demonstrating that when in need, promoters can rapidly emerge de novo.
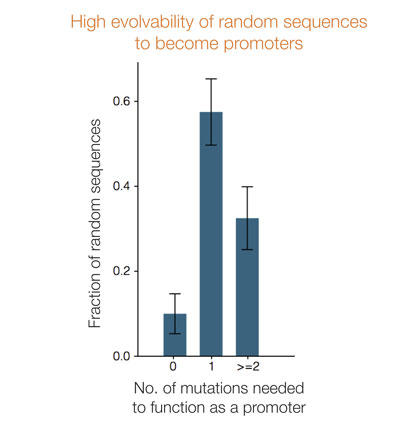
Figure: The promoter recognition machinery (RNA polymerase) was tuned to be one mutation away from recognizing any 100bp sequence as a promoter.
To mimic an ecological scenario that requires E. coli to obtain new promoters, Yona et. al deleted the natural promoter in E. coli that controls the expression of genes needed for the utilization of the sugar lactose as an energy source. The deleted promoter was then replaced by random sequences of the same size that were generated by the computer to have equal proportions of A, C, G and T. Now, with random sequences instead of a promoter, the cells have to find a new promoter otherwise they won’t be able to exploit the lactose in their environment.
To allow cells to search for a solution to this challenge, Yona and colleagues evolved the cells in a medium that is rich in lactose alongside little amounts of another sugar that the cells could use in order to survive. In this setup, any cell that finds a mutation that allows transcription of the lactose genes would have a fitness advantage because it can exploit the lactose in the medium without competitors.
Surprisingly, out of dozens of different random sequences the authors tested, 10% were already functional promoters that expressed the lactose genes (without evolution), 60% needed only one mutation to function as promoters, and the remaining 30% of the random sequences acquired a promoter by copying an existing promoter from other genes in the genome. These observations demonstrate how nonspecific sequences can easily turn into promoters in bacteria, even though they don’t contain any information – highlighting the incredible ability of biological systems to generate information out of chaos.
Why would evolution select for a promiscuous recognition system for promoters, where nearly any sequence is one mutation away from functioning as a promoter, and some are already promoters without the need for a mutation at all? The authors say that such a flexible system might be very beneficial for high evolvability of the transcriptional network. A system like this could help turn on new genes quickly – whether latent in the genome or taken in from other species through horizontal gene transfer.
However, this system may also lead to spurious transcription of unwanted fragments all across the genome. Interestingly, Yona et al. found that the E. coli genome was subjected to purifying selection that minimized the occurrences of “accidental promoters”, especially in cases where they are harmful, like interfering with the expression of essential genes.
This study also helps to explain the vast extent of horizontal gene transfer in the bacterial world. When bacteria adopt new genes from other species they have to be able to “read” the new genes. Since all forms of life share the same genetic code, any gene from any species maintains its meaning in the recipient bacterium. However, the gene’s regulatory sequences, like its promoter, might not be recognizable by the recipient transcription machinery. Yona et al. demonstrate that gaining transcriptional capacity of new genes is not a difficult step at all, hence increasing the fluidity of genes’ “exchange market” among different species.
Yona et. al. suggest a general design principle that might accelerate evolution of new features - on the one hand the specificity threshold for activity is set to be very low, almost on the verge of false activity. On the other hand, harmful “false positive” instances are deactivated. These two processes together accelerate evolution by creating a pool of functional (or almost functional) instances on which evolution can act by selecting beneficial instances. In this way, cells can enjoy high flexibility without paying a huge price for the low specificity.
The authors say that the need to balance between flexibility and specificity is so fundamental to biological processes that they wouldn’t be surprised if it holds true all the way to human cells. Therefore, one should expect some accidental transcription for human cells too. This perhaps can explain some of the unspecific activities along the human genome controversially interpreted as functional in the ENCODE project (a research project that aims to identify functional elements in the human genome). Furthermore, it could have lessons not only for transcription, but also for many other processes – from how enzymes recognize their molecular targets, and how proteins interact with other proteins - where you want to have this kind of molecular recognition.
As for his next step, Yona is planning to further evolve the promoters obtained from the current experiment so they will only be active when lactose is present. Since lactose was always present during the current evolution experiment, the evolved promoters exhibited constitutive expression, i.e., they were always on, even when we tested them on other sugars. Next, he plans an evolutionary regime that might select for promoters that only turn on when lactose is present, similar to the behavior of the natural lac promoter that he has deleted.
Avihu Yona, an HFSP fellow, says that the support from the Human Frontier Science Program for this project was crucial. “When I just suggested the project, my hosting lab thought that promoters cannot evolve from scratch, because they are too complex. They predicted that it is more likely for existing promoters to be copied into the random sequences. Nevertheless, when the first results emerged from the evolution experiments, we were all excited...”
Reference
Random sequences rapidly evolve into de novo promoters. Yona, A.H., Alm, E.J. and Gore, J., 2018. Nature communications, 9(1), p.1530.


































