Cells can tell the time. Because chemical reactions happen at certain rates, networks of these reactions inside cells act as timers, speedometers, and clocks. Telling the time is extremely important for cells, especially in developing organisms, where a tightly choreographed dance directs each cell to the right place at the right time.
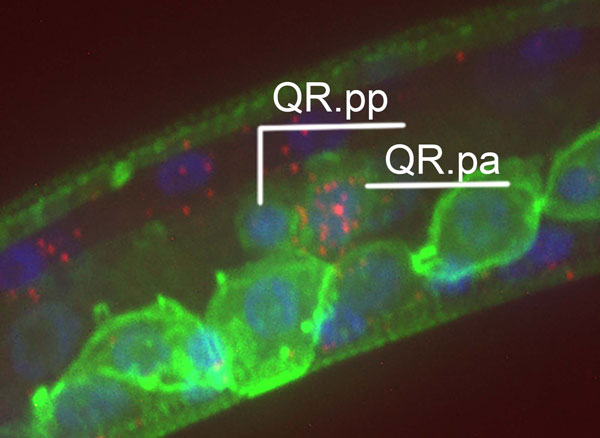
Figure: Single molecule fluorescent in situ hybridization visualization of mig-1 mRNA molecules in red. QR.pa shows strong, tightly controlled upregulation of mig-1 expression, while its sister QR.pp does not.
The simplest cellular timers involve a gene producing a protein and the cell detecting the time at which the amount of that protein crosses a particular threshold. However, even this simple timer is very noisy: there is a lot of randomness in the time it takes each protein molecule to be produced, and the threshold-crossing time can vary widely from cell to cell. To combat this noise, cells have evolved gene networks: one gene’s protein controls the protein production of another gene, which controls another, and so on. Depending on the architecture of these networks, they can reduce or enhance timing noise.
Particular network features, such as regulation chains, are known to decrease timing noise. Others, such as self-feedback, are known to increase noise. Surprisingly, we found using a mathematical model that combining these two opposing features decreases the timing noise below that of either feature individually. More generally, our theory predicts which specific network architectures decrease timing noise the most.
One of the optimal network architectures predicted by the theory appears to be at play in cells that migrate within developing larvae of the small worm Caenorhabditis elegans. The accumulation of a particular protein governs when the cell stops migrating, and this protein is subject to both regulation and feedback (the two key features of the theory). Our results suggest that the control of this protein may have evolved to optimize its timing precision. Our team is combining mathematical modeling with genetics and evolution experiments to unravel the choreography of organismal development.
HFSP funding was critical in developing our collaboration and getting this project off the ground. The PIs would not have met and begun working together if not motivated by the unique opportunity presented by HFSP.


































