One of the remarkable features of biology is that a cell can keep its extremely crowded proteome folded and organized without uncontrollable aggregation. Proteins are particularly prone to misfolding and aggregation relative to other classes of biomolecules because upon synthesis they typically require a complex folding pathway into a unique conformation; a process involving the shielding of hydrophobic moieties from aggregation. To govern folding and prevent aggregation, cells employ quality control networks comprising hundreds of proteins. A key role of these networks is to transiently engage with nascent proteins as they emerge from the ribosome, keep them folded, and degrade them should folding fail or when they reach the end of their natural lifespan. The net result is “proteostasis” (protein homeostasis), whereby the crowded proteome remains soluble and functional.
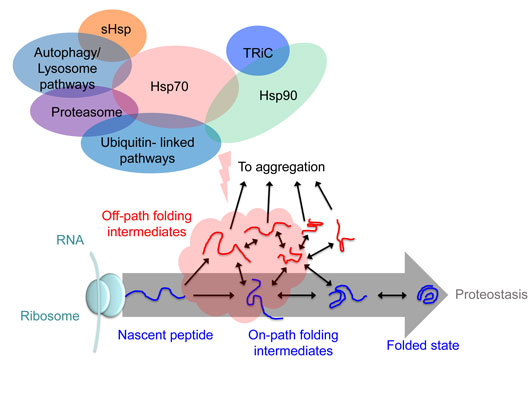
Figure: Conceptual framework for the biosensor. A network of proteins (some key families shown) form a quality control network to manage proteostasis. The proteins in this network can engage with different conformations of newly synthesized protein substrate and facilitate the efficiency of the folding reaction. In this example, Hsp70 family proteins can target unfolded proteins to foster correct folding and prevent their aggregation.
Quantitatively measuring how well the networks maintaining proteostasis operate remains a nebulous task. Proteostasis collapse or maladaptation leads to cellular malfunction and is associated with neurodegenerative disease. Proteostasis is also substantially remodelled to support cancer cell survival and changes (possibly detrimentally) upon normal aging. Hence, building capacity to measure proteostasis is important for understanding these maladies as well as understanding more fundamentally how cells manage the foldedness of the proteome under diverse conditions.
In this study, we describe a new strategy to quantitatively define changes in proteostasis balance. The approach involved construction of a new biosensor, which contained a metastably-folded protein (mutants of the prokaryotic ribonuclease barnase) that engages with chaperones and other quality control machinery. The biosensor readout relates to the changes in the available supply of quality control proteins. We used various approaches to positively and negatively influence chaperone supply to test and validate the biosensor strategy. A critical part of the project involved developing new approaches by flow cytometry to quickly and easily measure the biosensor readouts; namely how much chaperone bound to the biosensor and in the ability of proteostasis machinery to repress its aggregation as a metastable and hence aggregation prone protein.
The work involved important collaborations from members of our HFSP grant team. The Hatters lab members designed and built the biosensor system and validated it in cell culture models. The Ebbinghaus lab members characterized the dynamic range of the biosensor using in-cell thermal denaturation approaches and the Dickson lab devised the mathematical models to convert the biosensor readouts into mechanistic information. One of the interesting insights from the work was in applying the biosensor to define how heat shock protein 70 behaved in context of the intact cellular system. We found that the heat shock protein 70 chaperone cycle was rate limited by it binding to substrates under normal conditions, but this could be overcome by increasing levels of a nucleotide exchange factor (BAG1), which facilitates substrate release or alternatively by artificially activating the entire heat shock gene cluster by HSF1 overexpression. This indicates the potential of the biosensor to understand specific proteostasis machinery activity in context of an intact proteostasis system, as well as to report more generally on proteostasis health.
Reference
Biosensor-based framework to measure latent proteostasis capacity. Wood RJ, Ormsby AR, Radwan M, Cox D, Sharma A, Vöpel T, Ebbinghaus S, Oliveberg M, Reid GE, Dickson A, Hatters DM. Nat Commun. 2018 Jan 18;9(1):287. doi: 10.1038/s41467-017-02562-5.


































