RNA viruses are often considered to present the highest threat for triggering a worldwide pandemic. In the absence of effective therapeutic strategies, viral diseases such as dengue, Ebola, SARS and Zika exert a significant public health impact, while new RNA viruses continue to emerge, due to their rapid evolution. RNA viruses are so-called because they use RNA instead of DNA to represent their genetic code. This RNA genome codes for proteins and interacts with the host cell machinery. However, until now, the structure of viral RNA genomes while inside the host cell was largely unknown.
Artwork by Claudia Flandoli (http://www.claudiaflandoli.com).
The infection cycle of RNA viruses takes place mainly in the cell cytoplasm, where many host cell RNAs reside, whether these are messenger RNAs coding for proteins or non-coding RNAs that regulate diverse aspects of cell function. Virus and host RNA molecules can interact directly by base-pairing along parts of their structure. These interactions offer potential targets for anti-viral therapies, and indeed an anti-hepatitis C virus drug that targets such host–virus RNA interaction is currently in clinical trials. However, the prevalence of naturally occurring host–virus RNA base-pairing is unknown, and discoveries of new interactions are rare.
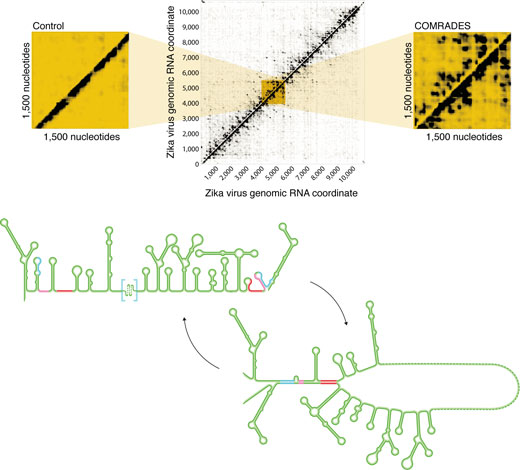
Figure: Exploring the structure of the Zika virus RNA genome inside living cells. Top: Crosslink Of Matched RNAs And Deep Sequencing (COMRADES) reveals the pattern of interactions between nucleotides along the viral genome, and a close-up shows the precision of the method. Each dot represents an interaction between the corresponding genome coordinates in the x and y axes. Bottom: Open and circular conformations of the viral RNA genome as computed from the resulting data. Figure by Claudia Flandoli (http://www.claudiaflandoli.com).
We have now developed a new technique to determine the dynamic structure of the Zika virus genome while replicating inside human cells as well as to identify critical base-pairing with the host RNA. The technique is called COMRADES (Crosslink Of Matched RNAs And Deep Sequencing) and, importantly, it can be applied to any RNA virus in any host cell. As RNA base-pairing can be dynamic, we employed small molecules named ‘cross-linkers’ that stably link the interacting partners to each other to provide a ‘snapshot’ of all RNA structures and interactions inside the cell. We revealed the first genome-wide structure of a viral RNA genome inside human cells and demonstrated that it is more dynamic than previously appreciated. We further identified multiple interactions with human regulatory non-coding RNAs such as micro RNAs, transfer RNAs and small nuclear RNAs.
As well as providing insight into how viruses direct the host cell to create new virus particles, the new method supplies the necessary and sufficient information for designing new strategies to interfere at specific interaction sites, thereby affecting the virus infectious cycle. The new method therefore opens the door to designing a new generation of RNA-based antiviral therapeutic approaches for a diverse range of RNA viruses. The support from HFSP was vital from initiation to completion of this research.
Reference
COMRADES determines in vivo RNA structures and interactions. Ziv O, Gabryelska MM, Lun ATL, Gebert LFR, Sheu-Gruttadauria J, Meredith LW, Liu ZY, Kwok CK, Qin CF, MacRae IJ, Goodfellow I, Marioni JC, Kudla G, Miska EA. Nat Methods. 2018 Sep 10. doi: 10.1038/s41592-018-0121-0. PubMed PMID: 30202058.


































