Cues from the environment and stresses can trigger inheritable physiological changes in our children and grandchildren. Such persisting traits or gene expression ‘memories’ do not involve alterations to DNA sequences, but instead are effected through a series of mechanisms termed epigenetics. The molecular mechanisms that underlie transgenerational epigenetic inheritance (TEI) have been difficult to trace over several lifespans in animal populations, or to perturb in a controlled manner. The short lifespan and experimental versatility of the nematode C. elegans makes it an ideal model to study these phenomena. In C. elegans TEI can be triggered and directed through the RNA interference (RNAi) pathways, which silences gene expression in response to endogenous or experimental double-stranded RNA sources. At the core of all RNAi pathways are the Argonaute proteins, which use small RNA molecules as a guide to direct gene-silencing activities. A subset of those Argonautes can enter the cell’s nucleus and instigate TEI in a pathway called the nuclear RNAi. Why and how some Argonautes do this was an outstanding question. The intersect between TEI and nuclear RNAi was the object of a HFSP project grant to the groups of Thomas F. Duchaine (Goodman Cancer Research Centre, McGill University, Canada), Eric Miska (University of Cambridge’s Gurdon Institute, UK), and Mihail Sarov (Max Planck Institute of Molecular Cell Biology and Genetics, Germany), together with collaborator James Wohlschlegel (UCLA, USA).
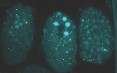
In a paper published in June in the journal Molecular Cell, the HFSP-funded collaborative revealed a family of proteins (ENRIs, for enhancers of nuclear RNAi) which control the strength and specificity of nuclear RNAi. Two ENRI proteins interact with the soma specific nuclear Argonaute NRDE-3. During embryo development they prevent loading of the Argonaute with the small RNA it uses as a guide to find its nuclear target transcripts (Figure 1). ‘A key finding was when we realized that the ENRI proteins specifically interact with Argonautes proteins that are not associated with small RNAs, or unloaded form’, says Alexandra Lewis, PhD candidate at McGill University and first author of the publication. Subcellular distribution and release of ENRI proteins is tightly restricted to specific stages of development and tissues, to ensure that Argonautes are activated at the right time and place (Figure 2). Upon loss of ENRIs in embryo, nuclear Argonaute NRDE-3 are aberrantly loaded with small RNAs that are reflective of the parental-identity, in turn leading to precociously translocation to nuclei. Loss of ENRIs further revealed a crucial function for nuclear RNAi in antagonizing activation of transposable elements. Reactivated transposable elements destabilize the genome, a striking phenotype named ‘mutator’. The collaborating HFSP grantees are now at work investigating molecular signaling events converging on ENRIs that may control their stability and sub-cellular localization.
These discoveries are transformative in the understanding of the biological purposes and the inner works of TEI and nuclear RNAi in development. They also unveil how animals acquire and regulate epigenetic memories against genome pathogens.
Figure 1 (appended as PDF file): ENRI-1/2/3 are small proteins that regulate the specificity of nuclear RNA interference by preventing the loading of Argonautes with small RNAs. Small RNAs guide Argonautes to their targets, which include deleterious pathogens such as transposons, and targeting by Argonautes triggers epigenetic silencing of targets. In the absence of such regulation in embryos lacking ENRI-1/2/3, the nuclear Argonaute NRDE-3 is precociously loaded with small RNAs which triggers unregulated and aberrant silencing.
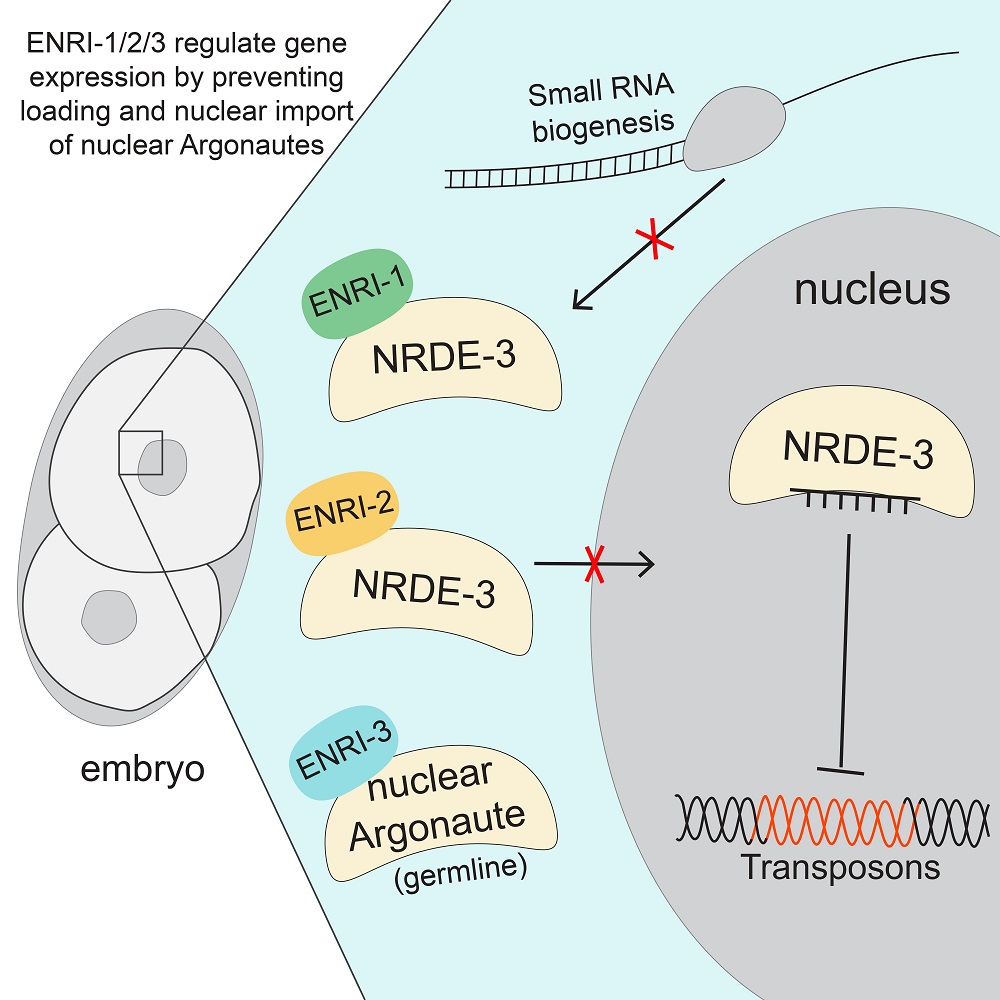
Figure 2: ENRI-2::GFP localization during embryonic development.Localization studies showed a dynamic behavior (nulear/cytoplasmic/condensate) of ENRI-2 during embryo development.