To survive and proliferate, living organisms must faithfully transmit their genetic information. However, modification of the DNA molecule is essential for organisms to adapt to new environments. This balance between genomic stability and plasticity is regulated by complex molecular networks known as DNA repair systems. Among the arsenal of mutagenic agents that can modify the chemical structure of DNA (oxidation, UV radiation...), alkylating agents are produced in many environments and attack nucleic acids of eukaryotic and prokaryotic cells. As a result, alkylating DNA damage repair systems are found in all living organisms.
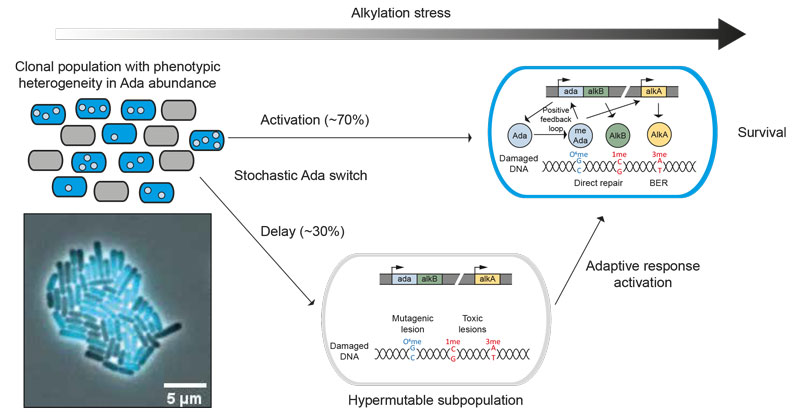
Figure: Cell-to-cell variability in DNA alkylation repair as a source of genetic plasticity. The stochastic expression of Ada splits an isogenic E. coli population into two distinct subpopulations. The subpopulation with a delayed adaptive response becomes hypermutable during alkylation stress. In contrast to a Δada-alkB hypermutable population, the delayed wild-type cells can eventually activate the adaptive response and thereby increase their chance of survival. This viable subpopulation can act as a source of genetic plasticity.
In E. coli, the response to alkylation stress, named the adaptive response, is composed of a network of three genes - ada, alkB and alkA - encoding DNA repair proteins with different enzymatic activities and substrate specificities. The expression of the adaptive response genes is induced by alkylation stress.
We visualized the timing of the adaptive response using a combination of single-molecule imaging and microfluidic-based single-cell microscopy. We found that the random activation of adaptive response genes generated a distinct subpopulation of cells in which all proteins of the adaptive response are essentially absent. As a consequence, this subpopulation is less efficient at repairing DNA alkylating lesions and accumulates more mutations. We propose that stochastic fluctuations of DNA repair capacity by the adaptive response creates a viable hypermutable subpopulation of cells that acts as a source of genetic diversity in a clonal population.
|
HFSP award information Long-Term Fellowship (LT000092/2019-L): Unraveling regulation of mutagenesis by DNA damage and antibiotic stress responses in single cells Fellow: Maxence Vincent |


































