 |
Takayasu Mikuni obtained his MD from Kyoto University. After which he worked as a pediatrician for five years. He then turned his focus to basic neuroscience, and studied for a PhD at the University of Tokyo. After postdoctoral training as an HFSP Long-Term Fellow at the Max Planck Florida Institute, he started his independent career as full Professor at the Brain Research Institute, Niigata University in Japan in 2018. His research is currently supported by an HFSP Career Development Award that he received in 2019. |
Thousands of proteins are known to mediate functional processes in a cell. Thus, precise, rapid and comprehensive mapping of subcellular localization of endogenous proteins is essential to describe cellular processes at the molecular level. However, conventional techniques have not provided rapid, scalable and high-throughput readouts for the localization of endogenous proteins, especially in the mammalian brain, due to the extremely high density and complexity of cellular processes.
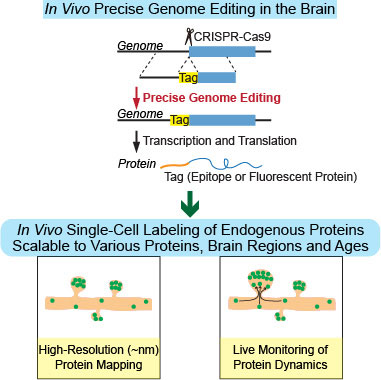
Figure: Strategy and application of SLENDR.
Direct, single-cell modification of the genome in vivo to insert a tag sequence to a gene of interest would provide rapid, specific and sparse labeling of the gene product protein. Using this strategy, we developed a simple and generalizable technique to image endogenous proteins with high specificity, resolution and contrast in single cells in mammalian brain tissue. The technique, termed SLENDR (single-cell labeling of endogenous proteins by CRISPR-Cas9-mediated homology-directed repair), enables in vivo precise genome editing to insert a sequence encoding an epitope tag or a fluorescent protein into a gene of interest. Although precise genome editing has not been possible in the mammalian brain in vivo due to the lack of homologous recombination activity in post mitotic neurons, we achieved single-cell, precise genome editing by delivering the editing machinery to dividing neuronal progenitors through in utero electroporation. We demonstrated that SLENDR is scalable to many species of proteins in diverse cell types and ages, and permits high-resolution imaging with light and electron microscopy both in fixed and live brain tissue.
Therefore, SLENDR allows researchers to rapidly and precisely determine the localization and dynamics of endogenous proteins with the resolution of micro- to nanometers in various cell types, regions and ages of the brain, providing a new platform suitable for large-scale analysis on a broad spectrum of proteins.


































