Animals and organisms often behave differently when exposed to a changing environment, and within an organism, even each cell can have unique behaviour. Among a population of cells with the same genotype there will be a range of responses and cells can choose different paths. Such differences in response can have wide effects ranging from development to ageing – from stem cells deciding whether to differentiate to cells deciding whether to either go through apoptosis or to selfishly transform into cancerous precursors.
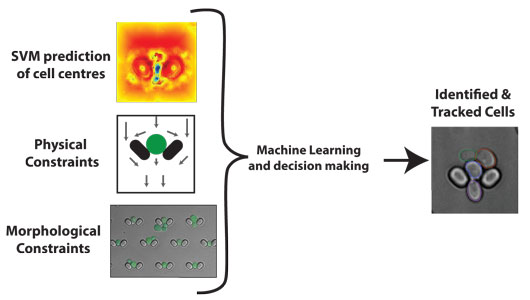
Figure: DISCO uses machine learning (a support vector machine) to find the centre of cells and combines this information with prior knowledge of both the design of the microfluidic imaging platform and of the changes in shape of yeast cells as they grow. By integrating these three approaches, DISCO robustly identifies and tracks cells over time.
To understand how populations of cells respond to environmental change and process information, many laboratories modulate the environment while imaging cellular responses using time-lapse microscopy. This procedure can generate terabytes of images that have to be processed and interpreted before providing any biological insight. Here we created a framework called Data Informed Segmentation of Cell Objects (DISCO) that incorporates knowledge about the shape of yeast cells, the study of which has given major insights into our own cells, to enable rapid and accurate automated image-processing.
For this framework we incorporated prior knowledge both about the size and shape of the budding yeast Saccharomyces cerevisiae and about the features of a microfluidic imaging platform. Prior work to automate the extraction of data from microscopy images has largely relied only on the images and finding characteristics to identify the cells. To improve upon these efforts, we used not just specific features from the images, but also our knowledge about the imaging platform employed, how this platform works, and how yeast cells grow and divide. By obtaining large numbers of quantitative measurements of cells, we generated a “shape space” that is used to determine the most likely outline of each cell. Similarly, by tracking and observing movement of cells within the microfluidic system we were able to improve predictions and tracking of individual cells from one image to the next in the time-lapse movie. As a result, we have already published novel insights into how biochemical networks have evolved to balance the competing needs of speed and accuracy when communicating information (1).
The computational framework is used by laboratories in both Europe and North America, and the HFSP funding was particularly instrumental in driving this intercontinental collaboration. As a result of the research resulting from this project, we expect additional publications on the consequences of genomic instability during mitosis, the costs of cellular misperception during aging, and how cells respond to environmental change.
- Distributing tasks via multiple input pathways increases cellular survival in stress. Alejandro A Granados, Matthew M. Crane, Luis F. Montano-Gutierrez, Reiko J. Tanaka, Margaritis Voliotis and Peter S. Swain, eLife, 6, e21415 (2017).
Reference
Morphologically Constrained and Data Informed Cell Segmentation of Budding Yeast. Elco Bakker, Peter S. Swain and Matthew M. Crane, Bioinformatics, 34 (1), 88-96 (2018).


































