Our understanding of complex biological systems often scales with the number of hypotheses we can test. Recent advances in high-throughput sequencing have allowed the testing of thousands to millions of different variants (DNA encoded hypotheses) at low cost using multiplex functional assays. Here variants are barcoded and tested together in a large pool. The input libraries to these assays have been limited to short DNA oligos of less than 200 bp (insufficient to encode a full length protein) or longer-length gene variants of a single particular sequence derived using mutagenesis approaches. The use of gene synthesis approaches has been limited in scale due to the large costs involved.
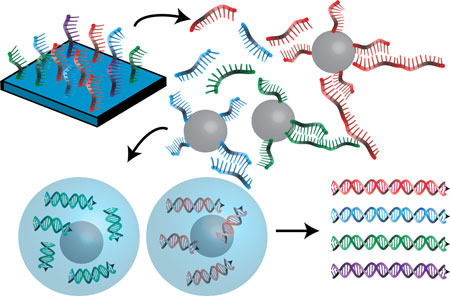
Figure: DropSynth uses barcoded microbeads to pull down the specific oligos needed to assemble a gene from a large oligo pool. Microbeads are then isolated within water-in-oil droplets, where the full-length genes are assembled using PCR. The assembled gene library is then recovered as a single mixture.
We have developed a novel method called DropSynth, which can assemble large libraries of genes at low cost. DropSynth leverages the cost and throughput of array-based oligo synthesis, in which thousands of oligos are made simultaneously in very small quantities. Each gene to be synthesized is split into a number of fragments with overlaps which can fit on a standard length oligo (~200 bp). Each oligo needed to assemble a particular gene is designed to contain a unique DNA barcode. These oligos are mixed with a large library of hundreds of barcoded beads, where each bead has a complementary barcode sequence meant to pull down all the oligos required for constructing an individual gene. After this multiplexed hybridization, the beads are vortexed together with oil to form an emulsion. This isolates the individual beads from one another inside water-in-oil droplets, preventing any interference during the assembly process and reducing costs drastically. Inside the droplets, the barcodes are enzymatically removed, and PCR is used to assemble the oligos into full-length genes.
We used DropSynth to assemble 12,672 genes encoding homologs of PPAT (coaD) and DHFR (folA), two ~500bp essential genes in the bacterium E. coli. We successfully assembled 7,143 genes (64% of verified genes) with a median of 1.9-3.9% perfects for each gene. DropSynth assembly costs around $1.50 per gene, far cheaper than other methods. The assembled PPAT libraries were used to test the ability of different homologs to complement a knockout E. coli strain in multiplex, revealing core functional motifs and reasons underlying homolog incompatibility, demonstrating the potential of this technique to functionally characterize thousands of genes. This approach could be used to screen for novel inhibitors against large libraries of genes encoding antibiotic targets.
Reference
Multiplexed gene synthesis in emulsions for exploring protein functional landscapes. Plesa C, Sidore AM, Lubock NB, Zhang D, Kosuri S. Science. 2018. 359 (6373), 343-347. doi:10.1126/science.aao5167.
DropSynth (resources for scientists interested in using the method)
Commentaries


































