Since Feynman’s 1959 lecture that introduced nanotechnologies as the means to leverage the properties of materials at the molecular level, there has been growing interest in the design of synthetic nanomachines. While the manipulation of molecules with atomic accuracy seemed improbable at the time, the engineering of molecular mechanical systems advanced considerably, albeit limited to synthetic chemistry. The design of dynamic protein mechanical systems is of great interest given their richer functionality: natural protein nanomachines evolved over billions of years to process energy and information in intricate ways by coupling biochemical energy to mechanical work, thereby supporting complex life forms. For instance, protein machines such as the ATP synthase are made of components resembling axles and rotor components capable of rotating relative to each other, which is leveraged by the cell to control the molecular energy supply. This suggests that artificial genetically encodable mechanical systems could be designed de novo to access a vast design landscape, by leveraging the sophistication of biopolymers organizational principles and sequence-based programmability. While recent advances in protein design now enable the generation of increasingly sophisticated static nanostructures and assemblies, the complex folding and diversity of non-covalent interactions has thus far made the design of dynamic protein assemblies very challenging.
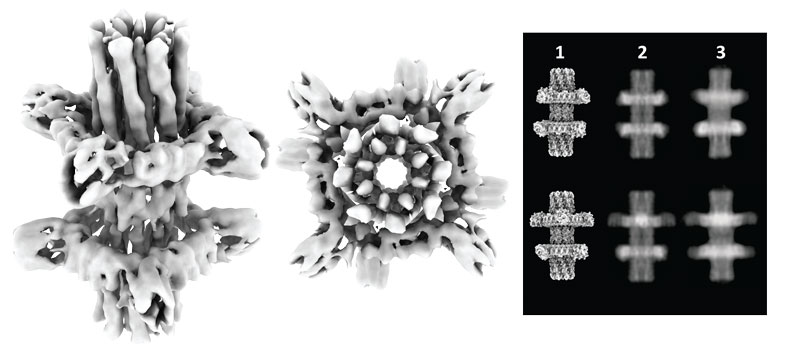
Figure: Overview of protein machine assembly. Artwork by ©Ian Haydon.
In this work, we leveraged the recent advances in computer-based design of de novo proteins, to take a step towards designing such nanomachines. We explored the computational design of protein mechanical systems through a first-principle, bottom-up approach that focuses on operational concepts independent from the complex evolutionary trajectory of natural nanomachines. We sought to design a nanoscale simple machine in which two mechanically coupled protein components undergo movement along chosen internal degrees of freedom. An important structural requirement to build such systems is that the interactions between components need to be strong enough to direct assembly while allowing rotational motion. We met this design challenge by computationally sculpting the energy of interacting components.
We report in Science that for the first time we were able to design rotary devices made from custom proteins. Such systems are made of nanoscale “axles” and “rotors” components designed to come together to form spinning assemblies, which are encoded in synthetic genes and introduced into living cells that carry out their fabrication. Using cryoelectron microscopy, we showed that the systems self-assembled and populated the mechanical degrees of freedom as designed. Such systems are a first step towards increasingly complex nanomachines. The capacity to accurately design the rotational energy landscapes is the basis towards the design of motor proteins, which could convert chemical energy into work at the nanoscale. This research paves the way for a new generation of nanoscale machines in which the mechanical motion is powered by solar energy or chemical fuels. One of our goals is to create nanomachines that could be used for medical or industrial applications, carrying out programmed complex tasks at the nanoscale, operating on cells or manipulating molecules in useful ways. We know by observing nature that very complex machines can be assembled from simple parts.
The project was led by synthetic biologist and biochemist Alexis Courbet, a postdoctoral scholar in the Baker lab and an HFSP Long-Term Fellow.
|
HFSP award information Long-Term Fellowship (LT000454/2016-L): A computational approach to designing self-assembling and programmable protein nanorobots Fellow: Alexis Courbet |


































