Building synthetic living cells by molecular assembly has become a major goal of constructive biology and biophysics. Among the different approaches to this ambitious goal, cell-free transcription-translation (TXTL) has established as an advantageous technology because it recapitulates gene expression in vitro. The archetype synthetic cell consists of a TXTL reaction encapsulated into cell-sized liposomes that are programmed genetically. Such a system enables biological functions to be isolated and developed dynamically at the scale of real living cells and within the natural physical boundary of a phospholipid bilayer. These experimental settings are also favourable to determine some fundamental properties of living cells. The self-assembly of the cytoskeleton into mechanically active supramolecular structures is of particular interest because it is an essential function for cell shape, cell motility and cell division.
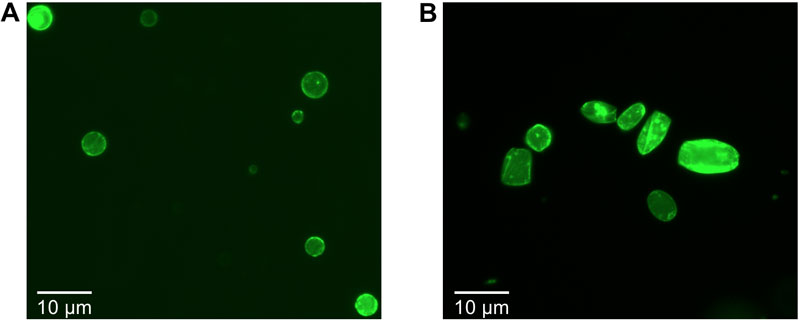
Figure 1. Cell-free synthesis of the MreB and eGFP-MreB proteins into cell-sized liposomes. The fluorescence images were acquired on an inverted microscope after 6 hours of incubation. A. Without the presence of molecular crowding at the membrane, MreB interacts with the lipid bilayer without inducing deformations of the liposomes. B. In the presence of molecular crowding at the membrane, MreB polymerizes into filaments capable of deforming spherical liposomes into rods.
In E. coli, the protein MreB polymerizes into filaments that shape the bacterium into a rod. Specifically, MreB interacts with the inner leaflet of the lipid membrane and self-assembles on the membrane into linear polymers. It is then capable of mechanically deforming the membrane into a rod, which also increases the physical stiffness of the microorganism. Without MreB, E. coli cells are spherical.
When MreB is expressed into spherical synthetic cells, one can observe a net interaction of the protein with the membrane. No deformation of the liposomes is observed, however (Fig. 1A) [1]. A major difference between the membrane of living and synthetic cells is due to two-dimensional molecular crowding. The membrane of synthetic cells is typically deprived of proteins, as opposed to the membrane of living cells covered by hundreds of different types of proteins. The effects of two dimensional molecular crowding on the self-assembly of proteins at the membrane have not been studied in synthetic cells, whereas three dimensional molecular crowding is a well-known effect to enhance self-assembly in bulk in vitro reactions.
To emulate two dimensional molecular crowding at the membrane of synthetic cells, the HFSP funded team used synthetic lipids composed of a synthetic polymer (PEG: polyethylene glycol) attached to a natural phospholipid. The PEG polymers emulated crowding at the surface of the lipid bilayer. This lipid was added to the membrane of synthetic cells, and its concentration was varied. When dynamically expressed into the synthetic cells, MreB spontaneously self-assemble into filaments capable of deforming the spherical liposomes into rod-like shapes (Fig. 1B) [1]. Deformations are observed for liposomes smaller than 10 µm radii. The smaller the liposome radius, the larger the deformation. Such deformations are only observed above a specific size of the PEG moiety and for molar concentrations between 0.2-3 % in the membrane. It is also important to notice that when membrane proteins are used rather than PEG, similar deformations are observed [2]. For all cases the deformations occur a few hours after the synthetic cells are prepared.
This study demonstrates that the self-assembly of MreB into a mechanically active cytoskeleton is driven by molecular crowding in two dimensions. We anticipate that achieving this symmetry breaking could lead to the development of other essential functions toward self-reproduction. The HFSP grant was truly essential to develop this research.


































