To study how transcription factors bind nucleosome-occupied motifs, we focused on the reprogramming factors OCT4 and SOX2. We determined cryo-EM structures of the pair of pioneering transcription factors bound to DNA on a nucleosome. One of the structures reveals how, by recognizing just part of its DNA-binding motif, one of OCT4’s DNA-binding domains appears to hold the DNA, while SOX2 lifts the DNA away from the surface of the nucleosome. This may indicate how the transcription factors open chromatin and allow recruitment of further factors for DNA transcription and subsequent gene expression. Using functional genomics approaches, we also showed that partial DNA motif engagement observed in the cryo-EM structure is sufficient in vivo for TF binding.
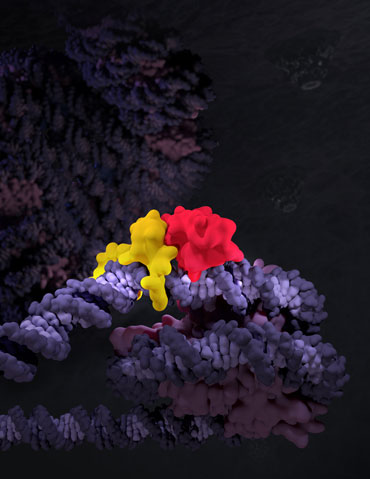
Figure: Reprogramming factors OCT4 and SOX2 pioneer chromatin. OCT4 (red) and SOX2 (yellow) are shown bound to a mononucleosome in the context of a chromatin fiber.
In order to further dissect the molecular engagement of pioneer transcription factors with nucleosomes, we developed an assay which showed us where the preferable sites for the transcription factor DNA binding motif on the nucleosome were, so that they could build transcription factor-nucleosome complexes. We hope that these techniques can be used to characterize the nucleosome-transcription factor binding relationship for other transcription factors in the future, and give further insight into how other necessary factors gather at open chromatin to initiate transcription and ultimately express genes.
HFSP funding was instrumental in supporting the first co-author Dr. Alicia Michael in this work now published in Science. In particular, the funding enabled Alicia to initiate collaborations leading to key experimental findings and supported attendance at international conferences to disseminate the work.


































