CRISPR/Cas9 gene engineering has revolutionized biology, however existing approaches have their limitations. While CRISPR/Cas9 gene editing is widely used in T cells, all described techniques require some form of culture or activation in a dish to make cells permissive to subsequent gene editing. This is a major stumbling block that limits the applicability of this technique, as culture can alter the way cells subsequently behave when transferred into whole animals. This is particularly problematic in the study of T cell tolerance, a poorly understood but fundamentally important immune process that helps to prevent the immune system from attacking “self” tissues. Prior culture can completely disrupt subsequent T cell tolerance, as the initial signals a T cell receives when exiting quiescence are critical to whether or not tolerance occurs. This has prevented the use of gene editing approaches in the study of tolerance within animal models, which has severely limited the field.
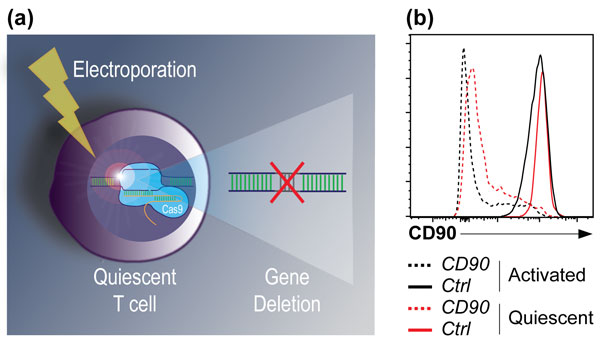
Figure: Effective gene editing in uncultured T cells expands the CRISPR/Cas9 toolkit to address unresolved immune questions in a fast-paced manner. (a) Schematic of the CRISPR approach used, and (b) CRISPR/Cas9-mediated deletion of the surface molecule CD90 in quiescent T cells (red lines) or T cells that were activated in a viral mouse infection model (black lines). Solid lines indicate cells given a control (Ctrl) sgRNA, while dotted lines are cells given a Cd90 sgRNA. Editing is comparable in both contexts, indicating that T cell activation is not required for gene deletion.
In the paper by Nüssing et al, which was funded by an HFSP Young Investigator Grant and recently published in the Journal of Immunology, the authors pioneer a highly efficient, electroporation-based CRISPR/Cas9-mediated gene editing approach that operates in uncultured, quiescent T cells. Using this method, they were not only able to delete genes in quiescent (or naïve) T cells, but further site-specifically edit a genomic locus, forcing the cells to express an altered surface molecule. These findings are surprising, as naïve T cells were assumed to be too metabolically and proliferatively quiescent to undergo either gene deletion, or the “homology-directed repair” process required for targeted gene editing.
Overall, these findings now open up a new gene editing toolkit for the study of T cell tolerance, a process fundamental to one of the central but incompletely understood features of the immune system: its capacity to discriminate between “self” and “foreign”. This approach is additionally of great utility when studying other immunological processes disrupted by T cell culture and activation, such as the pathways responsible for the survival and homeostasis of naïve T cells. Finally, this approach may be suitable for other quiescent and non-proliferative cell types that were previously assumed to be refractory to CRISPR-based gene engineering.


































