Several fluorescence microscopy methods can be used to measure protein interactions in cells. However, most of the methods struggle to capture both the diversity of these interactions across the entire cellular area and their dynamics over time. To overcome these limitations, we enhanced the spatiotemporal resolution of Number and Brightness, a spectroscopy method that transforms fluorescence fluctuations into protein aggregation values. The Enhanced Number and Brightness method implements two algorithms, the first one uses statistical tools to extract the distribution of protein oligomers (monomers, dimers, trimers and so on) in single pixels, thus revealing protein diversity across an entire cell image. The second one corrects the fluorescence intensity loss due to continuous light exposure, which enables measurements over prolonged periods of time. By combining these two algorithms, Enhanced Number and Brightness allows extended live imaging, providing dynamic maps of protein oligomerization overlaid on top of the corresponding cell image [1].
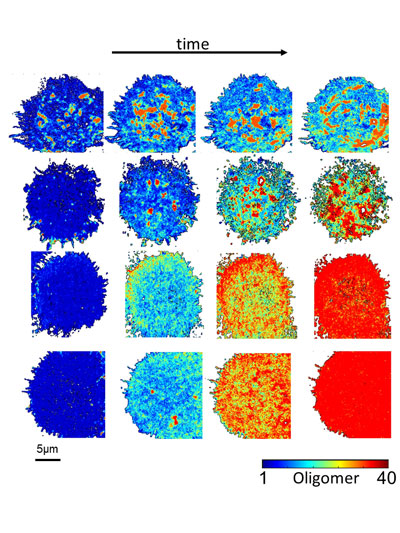
Figure 1. Enhanced Number and Brightness provides oligomerization maps (color-coded according to reference bar) during time-lapse movies. The image shows from left to right the dynamics of Eph receptor aggregation after stimulation with the ligand ephrin. Each row depicts a cell stimulated with different types of ligands (i.e. soluble, surface-bound etc.).
As a proof-of-concept, we recorded the activation of the Eph receptor, a key mediator of cell-to-cell communication [2]. The Eph receptor binds to its ligand ephrin, triggering the assembly of large receptor aggregates and the activation of the receptor by phosphorylation. The Enhanced Number and Brightness analysis of Eph clustering revealed that activation proceeds as a two-step process termed polymerization-condensation. During polymerization, small oligomers assemble leading to receptor activation. During the condensation phase, far larger aggregates form by the assembly of pre-activated oligomers, leading to endocytosis and signal termination. This dual mode of activation allows signal amplification (polymerization) and signal adaptation (condensation). This two-step process ensures high receptor sensitivity while keeping the dynamic range in check.
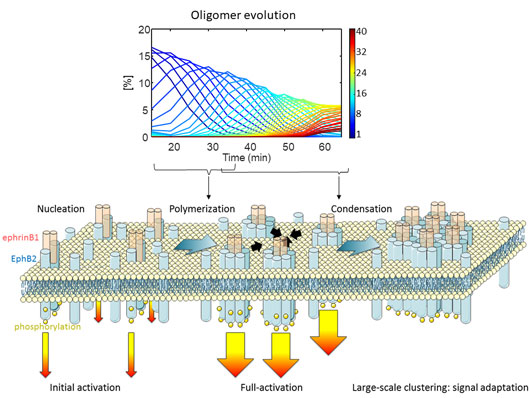
Figure 2. Top Panel, relative concentration of Eph receptor oligomers (color-coded) over time as retrieved by Enhanced Number and Brightness. Bottom panel, polymerization-condensation model of Eph receptor activation.
The Enhanced Number and Brightness resolution power proved to be high enough to discriminate among different modes of activation. We created ephrin nanopatterns of regular size (<30 nm in diameter) which induced a stronger receptor response than randomly distributed ligands, despite presenting a 9-fold lower ligand density [3]. The stronger, faster, and more efficient receptor activation of the nanostructured ligands provides a useful strategy to precisely tune and potentiate receptor responses.


































