The immune system is composed of many cell types that can detect the presence of invading pathogens and differentiate into effector cells with the capacity to eliminate harmful invaders. To mount an effective immune response and establish long lasting protection by generating an immunological memory, different types of immune cells must interact with each other and exchange signals. These include secreted molecules such as cytokines and chemoattractants as well as surface bound molecules that depend on cell-cell contacts in order to deliver a signal. These events take place within immune cell niches, small areas with lymphoid organs and other tissues where different immune cells congregate and exchange signals1. Many studies examine immune cell niches by immunohistochemistry that combines fluorescent antibodies labeling, genetically encoded fluorescent proteins and fluorescence based microscopy. However, these approaches are limited to a few markers that define a limited number of cells and do not allow analysis of gene expression.
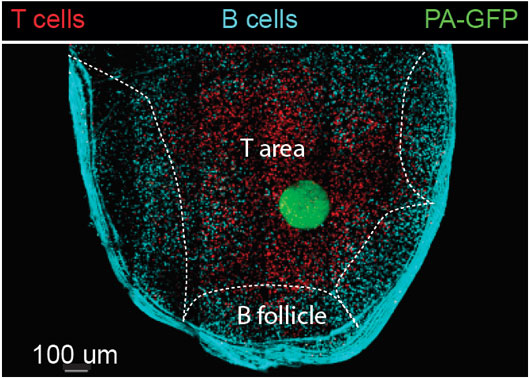
Figure: An image taken by a two-photon laser microscope of a lymph node with landmarks (T and B cells) that define different zones. An area within the T cell zone was photoactivated (green). The green cells (cells that originated from the T zone) were sorted and subjected to single cell RNA sequencing.
Single cell RNA sequencing is a technique that allows to determine the identity of single cells as well as their activation status based on their transcriptome2. However, this method lacks spatial information, and cannot determine whether a specific cell was present in a specific niche within an organ. PAGFP is fluorescent protein that upon exposure to UV light changes its emission wave length3,4. In order to examine the transcriptome of immune cells that were recovered from a known niche, mice that express PAGFP in all of their immune cells were used3. To define a niche, fluorescent cells were transferred to host PAGFP mice as landmarks of the area of interest. This specific area was exposed to UV light by two-photon laser scanning microscopy, activating especially the PAGFP in immune cells that reside within this zone. The labeled cells were sorted and subjected to single cell RNA sequencing. This technique was termed – NICHE-seq5.
By using this technique, the cellular compositions of various immune niches were defined. NICHE-seq analysis revealed that the area that filters the blood in the spleen is enriched with a special population of phagocytic cells. Furthermore, we found that upon viral infection a special type of immune cells (known as NK cells) accumulate in an area within the spleen where antibody producing cells (B cells) reside. Furthermore, in mice that were infected with fluorescently labeled virus, it was found that phagocytes are recruited to sites of viral infection and replication by upregulation of inflammatory chemoattractant receptors.
Collectively, NICHE-seq is a novel technique that combines information about the cell transcriptome and its location within an organ. This method is useful for reconstruction of any type of niche, including areas within the tumor microenvironment and discovery of molecules that mediate cell-cell communication.
References
1. Spatiotemporal Basis of Innate and Adaptive Immunity in Secondary Lymphoid Tissue. Qi, H., Kastenmüller, W. & Germain, R. N. Annu. Rev. Cell Dev. Biol. 30, 141–167 (2014).
2. Massively parallel single cell RNA-Seq for marker-free decomposition of tissues into cell types. Jaitin, D. A. et al. Science (80-. ). 343, 776–779 (2014).
3. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Victora, G. D. et al. Cell 143, 592–605 (2010).
4. Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science Shulman, Z. et al. (80-. ). 345, 1058–1062 (2014).
5. Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Medaglia, C. et al. Science (80-. ). 4277, eaao4277 (2017).
Featured in Research Highlights Nature Methods


































