The Mediator system is conserved across the eukaryotic kingdom and is required for the majority of protein coding gene expression. In yeast, Mediator comprises 25 subunits with a total mass of ~1.4 MDa and is organized into four modules, called head, middle, tail and kinase. The head and middle modules form the essential cMed, whereas the tail and kinase modules play regulatory roles. Mediator dependent phosphorylation stimulation of CTD by TFIIH is also important for capping, cleavage and polyadenylation of nascent transcripts. The mechanism by which Mediator enables regulated transcription is poorly understood, mainly due to the limited structural information.
Click on image to enlarge.
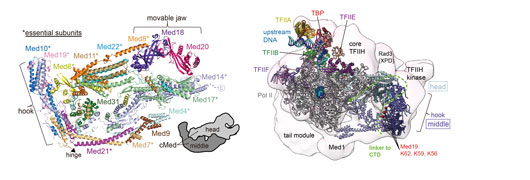
Figure: Ribbon model of the cMed structure refined at 3.4 Å resolution with the 15 subunits shown in different colours (Left). Model of the S. cerevisiae transcription initiation complex (Right).
In a long-term effort to solve the high-resolution Mediator structure, we co-expressed Mediator subunits in Escherichia coli using polycistronic expression vectors. We prepared recombinant cMed from fission yeast Schizosaccharomyces pombe, and improved protein solubility and yield by co-expression of Med1. Crystal dehydration improved the diffraction limit from ~8 to ~4 Å resolution and complete diffraction data to 3.4 Å resolution were obtained with a collimated synchrotron beam.
The obtained structure shows an unaltered head module compared to previous works, and newly reveals intricate middle module structure with five submodules: the beam, plank, hook, knob, and connector. The crystal structure also enabled us to obtain an atomic model of the Saccharomyces cerevisiae cMed structure. This structure informs us that sites of known Mediator mutations cluster at the interface between the head and middle modules.
Finally, the model for S. cerevisiae cMed could be combined with the 3.6 Å cryo-EM structure of the core PIC (cPIC) and the recently reported low-resolution cryo-EM envelope of a PIC–Mediator complex1, 2. The resulting atomic model comprises 35 polypeptides and includes Pol II, TBP, TFIIA, TFIIB, TFIIE, TFIIF, and cMed. This confirmed the proposed location of the tail module that had been predicted on the basis of superposition of free Mediator reconstructions. It also showed that the exposed end of the hook and the Med6 shoulder reached a density similar to that assigned to the TFIIH kinase subcomplex. The observed contacts between Mediator and TFIIH are probably relevant for Mediator-stimulated CTD phosphorylation by TFIIH. The CTD apparently extends from the body of Pol II along the Mediator head–middle interface into a previously described cradle. Superposition of the known head module–CTD peptide complex3 onto our cMed structure results in clashes of the modelled CTD peptide with Med4. Thus, the reported CTD positions or the cMed structure, or both, will change upon the PIC formation.
Reference
Core Mediator structure at 3.4 Å extends transcription initiation complex model. Kayo Nozawa, Thomas R Schneider, Patrick Cramer. Nature., 545, 248–251. doi: 10.1038/nature22328. Epub 2017 May 3.
Other references
1. Transcription initiation complex structures elucidate DNA opening. Plaschka, C., Hantsche, M., Dienemann, C. et al. : Nature, 533, 353-358, (2016)
2. Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Robinson, P. J., Trnka, MJ., Bushnell, DA. et al. : Cell, 166, 1411-1422 (2016)
3. Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Robinson, P. J., Bushnell, D. A., Trnka, M. J. : Proc Natl Acad Sci U S A, 109, 17931-17935 (2012)