Understanding which parts of the viral proteins can be targeted by the adaptive immune systems is a fundamental step in the development of vaccines. When viruses invade cells, small pieces of their proteins called peptides are presented on the cell surface by the human leukocyte antigen (HLA) complex to signal killer T cells. In turn, T cells eliminate the infected cells preventing the production and spread of thousands of new virions to neighboring cells. Although this process is critical in controlling viral infection and clearance, the repertoire of presented peptides remains unknown for many viruses including SARS-CoV-2, the virus causing the ongoing COVID-19 pandemic.
Reprinted from Weingarten-Gabbay et al., Cell. 2021 Jul 22; 184(15): 3962–3980.e17.
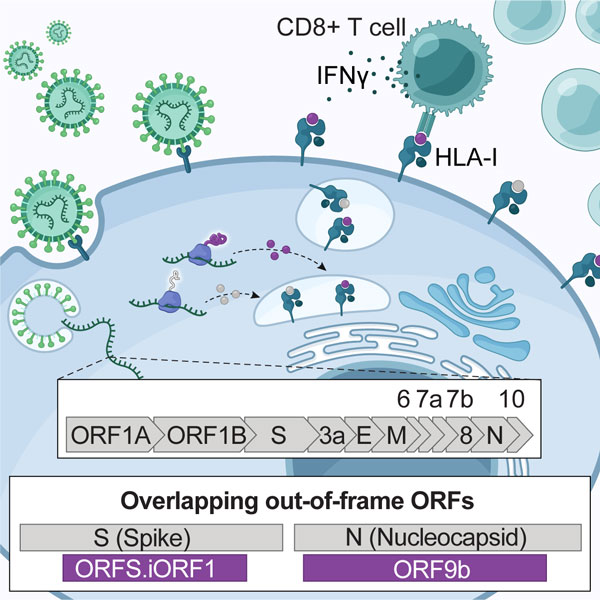
Traditionally, scientists who study the interaction between infected cells and T cells focus on the regions in the viral genome that encode the familiar viral proteins. Recent advances in sequencing-based technologies such as ribosome profiling, which allows high resolution measurement of translated RNAs, exposed the widespread existence of non-canonical open reading frames (ORFs) in ‘untranslated regions’ as well as overlapping reading frames. Employing ribosome profiling to cells infected with SARS-CoV-2 exposed 23 novel ORFs in addition to the 12 annotated ORFs (Finkel et al., Nature 2020).
While the function of these non-canonical ORFs in the viral life cycle is unknown, an HFSP postdoctoral fellow, Shira Weingarten-Gabbay, hypothesized that the translated polypeptides can enter the antigen presentation pathway and impact the immune response to SARS-CoV-2. In a recent study, Dr. Weingarten-Gabbay led a multi-institutional research collaboration aiming to provide an untargeted view of SARS-CoV-2 presented peptides using mass spectrometry (Weingarten-Gabbay et al., Cell 2021). Strikingly, the team found that 25% of the detected viral peptides originated from two non-canonical ORFs in the SARS-CoV-2 genome, suggesting that many potential targets for vaccines had been overlooked.
But does this hidden source of peptides signal T cells to attack virus-infected cells? To answer this question, the researchers probed the T cell responses to peptides derived from canonical and non-canonical ORFs in both humanized transgenic mice and blood samples from COVID-19 patients. They found that some of the non-canonical peptides provoked a stronger immune response than the familiar peptides. In fact, T cells from COVID-19 patients had greater responses to one of the non-canonical peptides than some of the most potent epitopes described in the literature to date. Gene expression analysis confirmed that the reactive T cells are memory cells, suggesting that they had ‘seen’ the non-canonical peptide in the past when the patients were infected with the virus.
The study sheds light on an important yet overlooked source for T cell targets that can potentially increase COVID-19 vaccine efficacy. Incorporating hidden T cell epitopes into future vaccines may improve T cell immunity. Perhaps more importantly, findings from this study uncover that common approaches in vaccine design may unknowingly destroy potent T cell epitopes from non-canonical ORFs. To increase the expression of a viral target, such as the Spike protein in the COVID-19 vaccines, scientists often change the native viral sequence in a process known as “human codon optimization”. Weingarten-Gabbay and colleagues demonstrate how these changes alter the amino acid sequence of overlapping non-canonical ORFs and eliminate T cell epitopes derived from these regions. This principle extends to other viruses as well, many of which utilize non-canonical ORFs to increase the coding capacity from relatively small genomes.
With the goal of exposing hidden T cell epitopes in hundreds of viruses pathogenic to humans, Dr. Weingarten-Gabbay develops innovative methods for the discovery of non-canonical ORFs in viral genomes as part of her HFSP supported research. She believes that a systematic view of viral antigen presentation, including non-canonical ORFs, will deepen our understanding of how viruses are ‘seen’ by T cells, provide new insights into viral immune evasion mechanisms, and allow the design of targeted vaccines.
|
HFSP award information Long-Term Fellowship (LT000396/2018-L): Deciphering viral immune evasion by systematic investigation of viral antigens |


































