We have shown that the self-assembly conditions of very simple peptide building blocks such as pH changes or alterations in the concentration of salts have a critical impact on the distribution of the corresponding supramolecular fibers on simple protocell models.
Spatial localization of cytoskeletal fiber polymerization is an essential factor that contributes to the successful emergence of cell dynamic properties, such as motility or cell division. For example, cytogenesis requires the exact positioning of the contractile ring in order to properly distribute chromosomes after mitosis. While the cellular machinery relies on a highly complex network of regulatory processes, our group asked the question of how simple parameters can determine this spatial localization on synthetic building blocks in order to provide a simple chemical approach for the emergence of these phenomena.
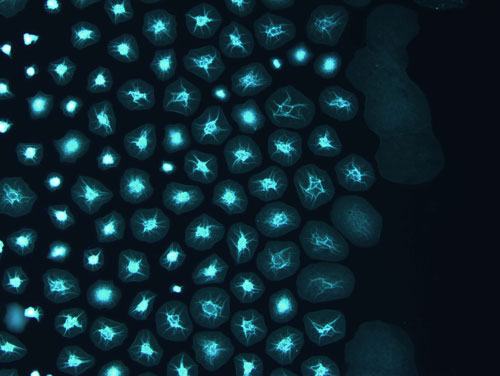
Figure: Synthetic self-assembled cyclic peptide fibrillar networks in water droplets, showing radial distribution in small spherical droplets and anisotropic growth in elongated droplets.
In our study we have used simplified containers, such us water-in-oil droplets containing simple synthetic building blocks (cyclic peptides), as a model for supramolecular fibers suspended in confinement. By using microfluidics, we have controlled, in a simultaneous manner, the generation of monodisperse, well-defined droplets and regulated the self-assembly of supramolecular fibers by adjusting the chemical environment (pH, ionic strength). We have shown that the assembly conditions provided a chemical basis for the specific localization of the supramolecular fibers on the droplet, allowing us to recreate cortex-like structures or dense-core networks. Remarkably, the microfluidic approach enabled fine-tuning of the spatial distribution of the microfibers’ networks. Furthermore, the spatial organization was exploited in the generation of 2D networks that inherited the droplets initial structure, yielding cortex and core networks, therefore expanding the organization by several scales.


































