Nucleic acids are one of the important building blocks of life. All living species on earth, including viruses, have their unique DNA or RNA genomic sequences and transmit the genomic information to next generations to enable them to thrive. Thus, analyses of genomic DNA/RNA sequences are critical for understanding how different forms of life work. DNA/RNA sequencing technology has been rapidly developed during the last two decades. Advanced sequencing technologies allow many researchers to produce diverse sets of DNA/RNA sequencing data from a variety of living species, including viruses, bacteria, animals and plants, and environmental samples. Even though these data are publicly available, it is not easy to search for specific DNA/RNA sequences of interest through all such datasets because of their huge size (individual datasets often include dozens of millions of sequence reads.) In this study, we developed a low-cost and high-speed tool for enabling meta-searches through a great variety of sequencing datasets.
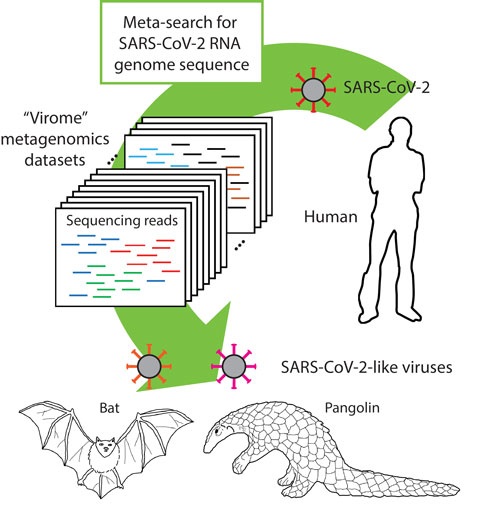
Figure: The distribution of SARS-CoV-2 RNA genome sequence in all available “virome” metagenomics datasets was assessed by using the meta-search tool that we developed. We detected sequencing reads that are highly similar to the SARS-CoV-2 RNA genome from bat and pangolin “virome” sequencing datasets. This figure was created by Min-Hye Kwon.
Part of my HFSP project was to apply efficient bioinformatic algorithms to facilitate sensitive and precise detection of rare transcripts from mRNA sequencing data. While we were developing the meta-search tool, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) had begun spreading all over the world and eventually caused the COVID-19 pandemic. To understand how the SARS-CoV-2 traversed the animal ecosphere, we explored the distribution of SARS-CoV-2 RNA genome sequences (refs 1-2) in all available “virome” metagenomics datasets. By using the meta-search tool that we developed, we searched for nucleic acid sequences that are highly similar to the SARS-CoV-2 RNA genome through the “virome” metagenomic datasets. We unexpectedly found SARS-CoV-2-like sequences in datasets from bat, bird, and pangolin meta-virome studies (refs 3-6). For sequence reads that matched the SARS-CoV-2 genome, sequences with the strongest and most abundant matches were detected in the datasets of pangolin lung meta-viromes. This observation raised a hypothesis that pangolins have been infected with SARS-CoV-2-like virus during the spread of the virus and in parallel to or connected to potential zoonosis to humans. At present it remains difficult to conclude, based on sequence analyses, whether pangolin contributes as an intermediate or secondary host to the evolution of human-infecting SARS-CoV-2.
Meta-searches of sequences of interest through big sequencing datasets provide opportunities to explore distribution of the DNA/RNA molecules in diverse environmental niches. The meta-search tool we developed in this study will be useful for tracing potential origins and the spreading of infectious agents like viruses or traversing transposable elements.
The support and flexibility of HFSP funding enabled (i) the development of novel and yet unknown meta-search tools, and (ii) the application of these tools to a set of questions that allowed us to investigate beyond the original proposal. The HFSP support opened an opportunity for making timely connections in a rapidly evolving and important research area that would otherwise have been challenging with standard task-based funding mechanisms.
|
HFSP award information Long-Term Fellowship (LT000329/2019-L) - Dissecting functional long non-coding RNAs and their working mechanisms |


































