Enzymes are tremendously proficient catalysts. They accelerate the timescales of biologically-relevant chemical reactions from millions of years (or more) to fractions of seconds. They are also “green catalysts”: environmentally-friendly, biodegradable, and with a low carbon footprint. These attributes make enzymes tremendously attractive as extracellular catalysts, to catalyze a whole host of industrially and pharmaceutically relevant natural and non-natural reactions, in particular in chemical industries where current catalysts are costly and with unfavorable environmental footprints. However, enzymes have had billions of years to evolve to operate under tightly regulated physiological conditions: how then do you (re)engineer their properties to be optimal under extra-cellular conditions?
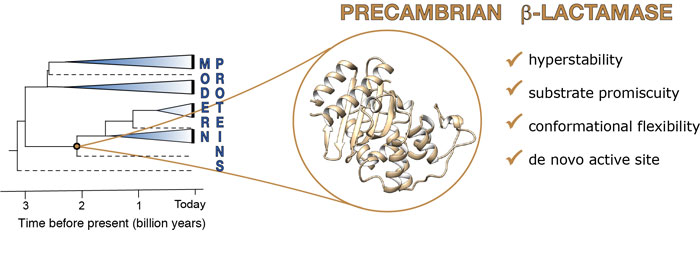
Figure: From ancient enzymes to modern activities: retracing billions of years of evolution to repurpose Precambrian enzymes for non-natural catalytic functions.
Recent years have observed two important advances in this direction: the first of these is the importance of conformational dynamics in enzyme evolution. That is, all enzymes are conformationally dynamic, in different ways, and on different timescales, from the conformational fluctuations of amino acid side chains in the active site through to large scale conformational changes that are necessary for functions such as loop or domain movements. While there has been substantial discussion of the potential catalytic role of such conformational changes, it is only recently that we have seen widespread acceptance of the evolutionary importance of such changes. It appears that Nature regulates conformational dynamics to facilitate the emergence of new functions – a feature that can be then mimicked and manipulated to reproduce the successes of Nature in the laboratory.
In parallel to this, a successful protein engineering venture is dependent on a high quality starting scaffold. So where to obtain such scaffolds? Most protein engineering effort has focused on either repurposing existing modern proteins, or on de novo design using computationally designed active sites grafted onto natural protein scaffolds. However, bioinformatics and biochemical techniques also allow us to perform ancestral sequence reconstruction, traveling back in time to predict the sequences of proteins that existed millions and billions of years ago, and to then reconstruct these proteins in the laboratory. These resurrected proteins typically demonstrate both higher thermostability than modern proteins, as well as high evolvability, making them excellent starting points for protein engineering.
In this work we discuss how the conformational dynamics of ancient proteins can be harnessed to repurpose ancient proteins towards modern catalytic activities, including completely non-natural reactions as was the case in ref. [1]. Here, we resurrected a β-lactamase enzyme from the Precambrian era and engineered it to catalyze a new reaction - Kemp elimination. This is a completely anthropogenic reaction that is frequently used as a benchmark system for enzyme design studies, among other reasons due to the fact that it provides an excellent model system for proton abstraction from carbon. Curiously, we observed proficient Kemp eliminase activity in the engineered active site of the ancient enzymes, and no activity in the modern enzymes. Computational and spectroscopic analysis suggested that this was due to significant differences in conformational flexibility between the different enzymes over evolutionary time. We then applied computational design to the most active of our repurposed ancient β-lactamases, in order to enhance the activity of this anthropogenic catalyst. Using a simple approach that combined evolutionary analysis of sequences with computational prediction of folding stability, FuncLib (http://www.funclib.weizmann.ac.il), we were able to obtain designed variants with catalytic efficiencies to match that of modern enzymes (ref [2]). This is particularly significant as FuncLib, being based on evolutionary analysis of sequence evolution, is primarily designed to target natural activities in natural enzymes. However, we demonstrate that also in the case of non-natural activities in de novo engineered active sites, FuncLib is a powerful tool for enzyme design.
We have also explored the feasibility of using ancestral TIM-barrel glycosidases as scaffolds for the design of enzymes with new catalytic properties. As part of this, we have recently resurrected an ancient TIM-barrel glycosidase with exceptional catalytic properties, and demonstrated that heme-binding allosterically regulates its catalytic activity [3]. This work was facilitated by an HFSP grant focusing on repurposing conformational dynamics to manipulate the catalytic activities of ancestral proteins.


































