SARS-CoV-2, the coronavirus causing the ongoing COVID-19 pandemic, infiltrates our cells with the help of the spike protein. Spike is present on the surface of the virus, giving it the characteristic crown-like appearance, and docks onto surface receptors on the host cell. In this study, Sikora and coworkers revealed how the spike protein evades the immune response and identifies weak points in its protective cloak [1].
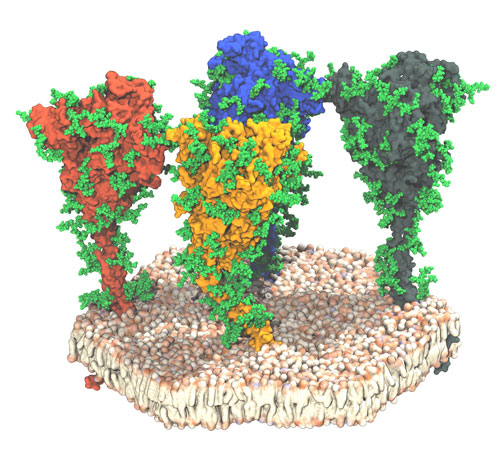
Figure: snapshot of a molecular dynamics simulation of four spike proteins (differently colored) embedded in a membrane. The glycan “cloak” protecting the spike protein against the immune system is shown in green. Credits for the figure: Mateusz Sikora, Sören von Bülow, Florian E. C. Blanc, Michael Gecht, Roberto Covino and Gerhard Hummer.
Last year, researchers at the Max Planck Institute of Biophysics (MPIBP), together with colleagues at EMBL and the Paul-Ehrlich Institute, published a study [2], supported by HFSP, showing that spike is a surprisingly flexible molecule, and that this flexibility could facilitate the viral entry into host cells. In a new study by Sikora and coworkers [1], the researchers at the MPIBP now focused on how spike hides from the watchful eyes of our immune system. When we get infected by a virus, our bodies produce antibodies that latch onto the virus and expose it to the machinery of our immune system. However, coronaviruses have developed a powerful 'invisibility cloak' to obfuscate our defenses: the spike protein is covered by highly dynamic sugar-like molecules called glycans that conceal its surface.
The researchers performed molecular dynamics simulations to study conformational changes in the spike protein and its glycan cloak. These simulations are powerful tools to study biomolecules at incredibly high spatial and time resolution. This way, Sikora and his team members found that the glycans sweep off incoming molecules on the spike surface, just like windshield wipers on a car. Even though at any given time only a small part of the spike surface is covered by the glycans, the rapid glycan motion effectively cloaks the protein from our immune system.
The scientists then set out to find holes in this invisibility cloak by identifying spots on the spike protein that are less effectively shielded by glycans. To this aim, they analyzed different features on the spike surface like accessibility, rigidity, and sequence variability. This analysis was sped up significantly by building on a technique originally developed in another collaborative study supported by HFSP [3] to analyze Atg9 vesicles implicated in autophagy. From the analysis, nine regions on the spike surface were identified as promising targets for the development of vaccines and therapeutics. The technique developed here may also prove useful in the study and treatment of virus variants and other viruses.


































