In recent years, organoids have increasingly been applied to developmental biology, biomedical and translational studies. Organoids display a high phenotypic complexity that is usually captured with a wide range of microscopy techniques and platforms, from common benchtop to high resolution imaging systems. To understand the underlying mechanisms responsible for organoid development, it is necessary to extract useful information from diverse microscopy images. However, because of their phenotypic complexity and variability, organoid studies focus on high-throughput, high-content microscopy, which generates large amounts of data that are becoming increasingly difficult to inspect and interpret.
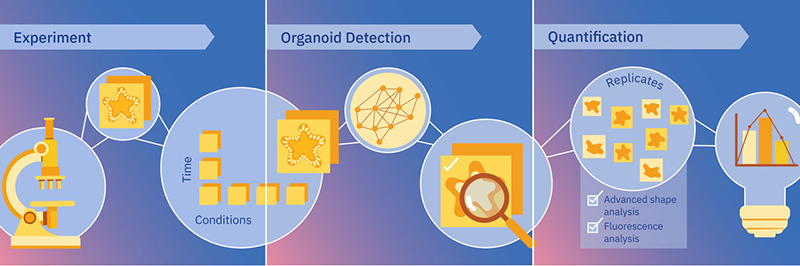
Figure: MOrgAna workflow from image acquisition, to organoid recognition and quantification. Credits: Rayne Zaayman-Gallant/EMBL.
An ideal way to extract information from large amounts of data is machine learning (ML), which is capable of learning non-trivial rules by iteratively inspecting and interrogating the data. In the microscopy image analysis field, ML is capable of extracting key features in an automated, reproducible and quantitative manner. However, ML algorithms generally require programming knowledge, and are therefore employed by only a few experts. As a consequence, little effort has been made to develop a pipeline that can be used by the large organoids community with limited programming experience.
Through this work, we introduce MOrgAna (Machine Learning-based Organoids Analysis), our solution to provide a user-friendly pipeline to analyze hundreds of organoids images. MOrgAna allows the end user to quickly run ML algorithms through an intuitive graphical interface and visualize the results with a simple click. Behind its pretty face, MOrgAna implements deep learning algorithms to classify pixels within an image into three types: background, organoid, and organoid edge. This is a particularly important feature, as organoids typically become surrounded by dying cells and debris during the course of their development, thus making it difficult to detect their boundaries accurately.
MOrgAna is not only capable of computing “simple” parameters such as the area and perimeter, but extends to more complex shape and fluorescence descriptors. One of them is LOCO-EFA (lobe contribution elliptic fourier transform), which is utilized to describe the number of protrusions, a hallmark of development in many types of organoids. Another example is the fluorescence intensity profile along a given organoid axes. This measure is used, for instance, to characterize the anterior and posterior parts of gastruloids, a type of organoid that displays axial organization, thus allowing researchers to study the earliest stages of mammal development in a dish.
Finally, it was important for us to provide a freely accessible tool, which we believe fosters interactions between scientists with different backgrounds, thus promoting open, collaborative and interdisciplinary research.
|
HFSP award information Long-Term Fellowship (LT000227/2018-L): Quantitative analysis of single-cell regeneration dynamics in living planarians Fellow: Nicola Gritti |


































