In metazoans, upon transcription of a gene into RNA, introns are removed but exons can be stitched back together in different ways in a process termed alternative splicing. The resulting alternative gene products can differ functionally, and regulated alternative splicing thus plays important roles in cell differentiation. While many splicing factors are known, including the components of the multi-subunit spliceosome, it is only recently becoming clear that chromatin-associated proteins impact alternative splicing as well.
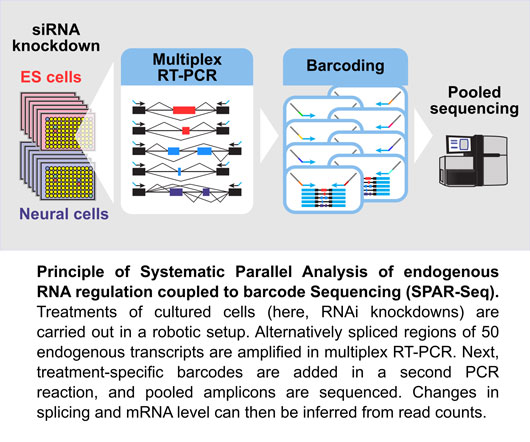
Han, Braunschweig et al. developed “Systematic Parallel Analysis of endogenous RNA regulation coupled to barcode Sequencing” (SPAR-Seq) as a flexible screening platform that allowed them to probe more than 1,400 potential trans-acting factors – known and putative RNA binding proteins as well as chromatin and transcription factors – for their impact on any of 50 endogenous splicing events with roles in cell identity (see Figure).
Screens in mouse embryonic stem and neural cells linked proteins involved in various aspects of the mRNA life cycle to alternative splicing regulation and highlighted physical and genetic interactions among members of protein complexes and regulatory pathways. Hundreds of proteins with no known roles in splicing were found to impact one or more of the interrogated alternative splicing events.
Surprisingly, these included many DNA-binding transcription factors. Further analyses showed that some transcription factors not only regulate the expression of splicing factors, but are also able to bind to mRNAs near regulated exons and thus contribute to the coordinated regulation of networks of alternative splicing events at multiple levels. Together with accumulating mass spectrometry-based evidence that a large number of proteins with no canonical RNA-binding domains bind to mRNA, these findings paint a picture in which different levels of gene expression regulation are more intricately interconnected than had previously been recognized.
Reference
Multilayered control of alternative splicing regulatory networks by transcription factors. Hong Han*, Ulrich Braunschweig*, Thomas Gonatopoulos-Pournatzis, Robert J. Weatheritt, Calley L. Hirsch, Kevin C.H. Ha, Ernest Radovani, Syed Nabeel-Shah, Tim Sterne-Weiler, Juli Wang, Dave O’Hanlon, Qun Pan, Debashish Ray, Hong Zheng, Frederick Vizeacoumar, Alessandro Datti, Lilia Magomedova, Carolyn L. Cummins, Timothy R. Hughes, Jack F. Greenblatt, Jeffrey L. Wrana, Jason Moffat, and Benjamin J. Blencowe. Mol Cell (2017), 65(3):539-553.. DOI: http://dx.doi.org/10.1016/j.molcel.2017.01.011 *equal contribution


































