Two types of methods are classically used to identify microRNA targets: molecular biology (e.g., immunoprecipitation followed by deep-sequencing to identify microRNA-interacting mRNAs) and bioinformatics (searching for phylogenetically conserved microRNA binding sites). Molecular biology methods can detect every sort of interaction, including those that do not play any biological role at the macroscopic scale. Indeed microRNAs repress most of their targets to a very small extent (less than 2-fold in general) whereas the biological activity of most genes is robust to such small fluctuations in expression.
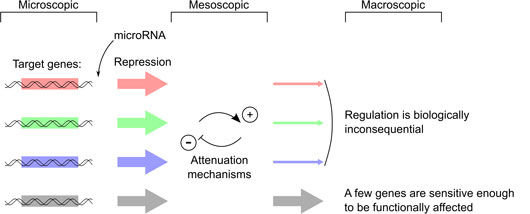
Figure: microRNA-mediated repression can be measured for thousands of targets at the microscopic scale. However, our results suggest that most of these molecular events are inconsequential at the macroscopic scale (they are attenuated by homeostatic mechanisms): only the most dose-sensitive genes can be functionally targeted.
Bioinformatics methods are supposed to exclude inconsequential molecular interactions: one usually considers that genomic elements are conserved only if they play a beneficial role at the organism scale. Therefore, the scientific community usually combines molecular biology and bioinformatics to identify molecular interactions that play an important biological role.
However, we found several sources of false positives in these methods. First, focusing on a neutrophil-specific microRNA, we observed that most computationally predicted targets appear to be functionally insensitive to the microRNA: inter-individual variability in gene expression among wild-type mice frequently exceeds microRNA-mediated regulation, while such variability does not trigger any macroscopic phenotype. Second, we identified two conceptual issues that contaminate current computational predictions with false positives:
- Some mRNAs are so abundant that they can sequester a large fraction of the microRNA pool. They can thus modulate microRNA availability for other targets. Such microRNA-modulating activity can constitute a biological function subjected to selective pressure. It is thus likely that some microRNA binding sites are conserved because of that novel function, rather than because of their weak effect on mRNA expression.
- Some genomic regions are conserved because of microRNA-independent reasons (e.g., protein binding) while being fortuitously complementary to a microRNA sequence. Computational programs consider that these sites are conserved because of microRNA binding, when they are not. We could estimate the number of such erroneous assumptions by counting microRNA binding sites that are more deeply conserved than the microRNA itself (e.g., a mammal-specific microRNA cannot be responsible for the conservation of a binding site that is conserved between mammals and fish).
Our work thus questions the function of many published microRNA/mRNA interactions. Among the hundreds of proposed targets for each microRNA, we propose that just a few of them (the most dose-sensitive genes) are real targets, in a biological sense – others would just be inconsequential molecular events.
Reference
The number of biologically relevant microRNA targets has been largely over-estimated. Natalia Pinzón, Blaise Li, Laura Martinez, Anna Sergeeva, Jessy Presumey, Florence Apparailly and Hervé Seitz. Genome Research, in press. doi : 10.1101/gr.205146.116.


































