Antibiotic or drug resistance of microbes has become a global concern in the past few decades. As resistance genes are commonly carried on plasmids, which are circular, self-replicating, mobile pieces of extra-chromosomal DNA, the maintenance of plasmids in bacteria over generations is of great importance. Particularly, the maintenance of smaller plasmids at high-copy-numbers is challenging, due to the lack of active segregation mechanisms, and relies significantly on their spatial organization inside bacteria, which was not clear.
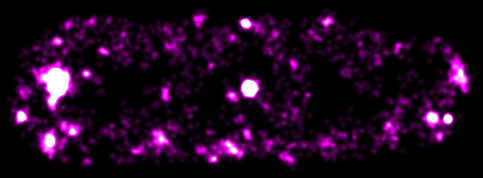
Figure: A super-resolved fluorescence image of ColE1-derivative plasmids in an E. coli bacterium.
HFSP Cross-Disciplinary Fellow Yong Wang and colleagues (Paul Penkul and Joshua N. Milstein) recently investigated the spatial organization of a high-copy-number plasmid in E. coli bacteria using quantitative localization microscopy combined with single-molecule fluorescence in situ hybridization. They observed two populations of the plasmids simultaneously present in the bacteria: some of the plasmids aggregated into large clusters, and many plasmids were randomly distributed along the axes of the bacteria. For the first time, both the clustering and non-clustering high-copy-number plasmids were detected, indicating that previous observations on the spatial distribution of plasmids in bacteria were incomplete. Furthermore, this work indicated that the non-clustering population of high-copy-number plasmids should be efficient enough for the maintenance of plasmids in bacteria over generations, and therefore alleviate the need of undiscovered active mechanisms.
In addition, this work showed that, although the plasmids were preferentially distributed at the periphery of the bacteria, likely due to steric exclusion by the chromosomal DNA comprising the nucleoid, the plasmids appeared through routes at the central region of individual bacteria. This observation implies that the excluded volume of the nucleoid is more amorphous than was previously thought.
Reference
Quantitative localization microscopy reveals a novel organization of a high-copy number plasmid. Wang Y, Penkul P, Milstein JN. Biophys. J. 2016 Aug;111(3):467–479. doi: 10.1016/j.bpj.2016.06.033.


































