Only 15 years ago, scientists discovered the ESCRT-III protein complex. This protein complex actually plays an essential role in the key moments of a cell’s life. This complex is behind the final phase of cell division, when the membrane is cut, allowing the daughter cells to divide. ESCRT-III also helps some viruses (such as HIV) to separate themselves from the host cell by cutting the virus bud attached to the cell membrane. More recently, it was found to seal holes left in the nucleus envelope after it was rebuilt at the end of cell division. ESCRT-III was originally discovered in the lysosome pathway, where organelle membranes are budded inwards to produce Intra-Luminal vesicles in the endosomes, that will then be degraded in the lysosomes. In this pathway, ESCRT-III has been implicated in both the deformation of the membrane and the fission (breaking of the neck). Interestingly, ESCRT-III has been implicated in most of the fission reactions where cytosolic proteins reach the lumen of the membrane neck to be cut. ESCRT-III is thus, probably, a general fission machinery for these reactions.
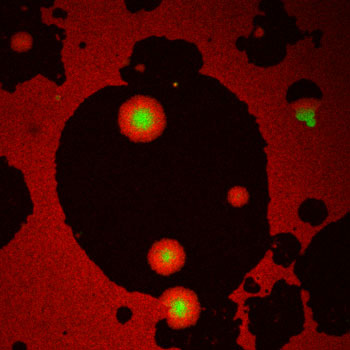
Figure: Protein patches made by the ESCRT-III protein Snf7 (green disks surrounded by red circular gradients) at the surface of lipid membrane (black shadows).
Like a watch spring
Researchers from the University of Geneva (UNIGE), INSERM (the French National Institute for Health and Medical Research)/Aix-Marseille University, and the French National Center for Scientific Research (CNRS) have just understood how ESCRT-III operates. Like a lego brick, the proteins fit into each other until they form a flat spiral at the surface of the membrane. To understand how these molecules form such spirals, Nicolas Chiaruttini, in Geneva, developed a fluorescent microscopy in vitro assay where the ESCRT-III protein Snf7 polymerized onto supported lipid bilayers by a nucleation growth process on a large scale (tens of microns): small nuclei appeared at a low frequency on the membrane, and then expanded into circular patches that grew and merged to cover the surface. To unravel the molecular structure of these patches, Simon Scheuring’s group looked at the polymerized Snf7 with Atomic Force Microscopy (AFM), and observed densely packed spirals that looked like spiral springs found in watches. Postulating that these spirals are springs that stop growing when laterally compressed, Martin Lenz built a theoretical model that could recapitulate the growth process at the macroscopic (micron) level. To validate the microscopic assumptions, the researchers took advantage of a remarkable technique (see below) called High-Speed AFM, which allows imaging of molecule movement at video rate. They observed growing Snf7 spirals, and showed that they are indeed highly flexible and compress. Further biophysical measurements allowed the rigidity of Snf7 filaments to be estimated, which was found to be 5-10 times smaller than actin, and polymerization energy, which was found to be relatively high, equivalent to that of clathrin. These values confirmed that Snf7 can be deformed by its own polymerization, and store enough energy in a single spiral to form a vesicle.
How would this spring behaviour deform the membrane? Polymerization into a spiral results from the properties of the Snf7 filament to curl at a given radius (25-30nm) and to be flexible. Snf7 initially forms rings of 25-30nm, which break, and stumps grow further into a spiral, because of the flexible nature of the Snf7 filament. Another consequence of this flexibility, is that as the spiral grows, inner filaments of the spiral are compressed, and outer ones are stretched, as they are not at their preferred radius. Similar to watching a spring being wound-up, over-compression accumulates the energy required to start the system. When the pressure is too high, the spring can “buckle”, which means that the inner filaments will go off the plane, to allow them to expand. In doing so, they will push and deform the membrane. This mechanism is similar to broken clocks, in which the spring is overloaded and breaks by pushing the frame out. This buckling mechanism could explain striking observations where plasma membrane tubules coated with ESCRT-III spirals have been observed in cells overexpressing ESCRT-III molecules (2, 3).
In this study, the authors did not observed buckling. However, they showed that releasing the compression of the spirals would roll the membrane, in a process known as “curling”. This effect came from the fact that when Snf7 spirals expanded, they created an area difference between the membrane leaflet they are bound to and the opposite leaflet. To accommodate this area difference, the membrane spontaneously curls, showing that Snf7 spiral spring expansion is strong enough to deform the membrane.
The latest technology
A combination of the skills of biochemists, physicists, and theoreticians was required to understand the molecular mechanics of this complex. The theoretical estimated stored energy and the spring strength that Martin Lenz from the CNRS estimated were validated by the biophysical experiments conducted in Geneva.
With the latest technology, the researchers were able to observe the movements of the complex in real time and at the nanometer level. This feat was achieved with an atomic force microscope (AFM), the only one of its kind that can immediately provide nanometric resolution, by the team of Simon Scheuring, INSERM Research Director, Aurélien Roux, a biochemistry professor at the UNIGE Faculty of Science, is pleased to say that this is the first time that this technique has been used for this kind of work, and that this proves, yet again, that interdisciplinary cooperation leads us on original paths.
Reference
Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, Scheuring S, Roux A. Cell. 2015 Nov 5;163(4):866-79. doi: 10.1016/j.cell.2015.10.017. Epub 2015 Oct 29. PubMed PMID: 26522593; PubMed Central PMCID: PMC4644223.
Other references
1. Membrane buckling induced by curved filaments. Lenz M, Crow DJ, Joanny JF. Phys Rev Lett. 2009 Jul 17;10(3):038101. Epub 2009 Jul 13. PubMed PMID: 19659322. (This is the theoretical paper (published in PRL) that proposed the spiral spring/buckling hypothesis.)
2. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. P. I. Hanson, R. Roth, Y. Lin, J. E. Heuser. The Journal of cell biology 180, 389 (Jan 28, 2008).
3. Structure and membrane remodeling activity of ESCRT-III helical polymers. J. McCullough et al. Science, (Dec 3, 2015).


































