The emergence of a complex eukaryotic cellular plan is one of the greatest evolutionary marvels. It is now believed that an archaeon engulfed a bacterium nearly 2 billion years ago, evolving into a stable endosymbiotic relationship that gave rise to the present-day eukaryotic cell body and its mitochondria. The discovery of Asgard archaea from environmental metagenomes has brought much excitement in the past few years as they were shown to be the closest archaeal relatives to the present-day eukaryotes. In recent phylogenomic analyses, one branch of Asgard archaea named Heimdallarchaeota has been indicated to be the closest to eukaryotes. Yet, their biology has only been inferred from a few fragmented environmental metagenome-assembled genomes (MAGs).
As part of an HFSP-funded study on the energy flow governing the carbon and nitrogen cycle, Wu and colleagues amended diverse energy sources to the rock and sediment samples retrieved near Diane’s vent in the 3.7-km-deep Southern Pescadero Basin (with its stunning image featured on the cover of Nature Microbiology, February 2022). Besides finding the roles of energy availability on deep-sea microbial carbon flux (Wu et al, in preparation), some energy sources caused a significant reduction in community complexity and the clonal expansion of Asgard archaea, bringing an unexpected advantage for metagenomic assembly.
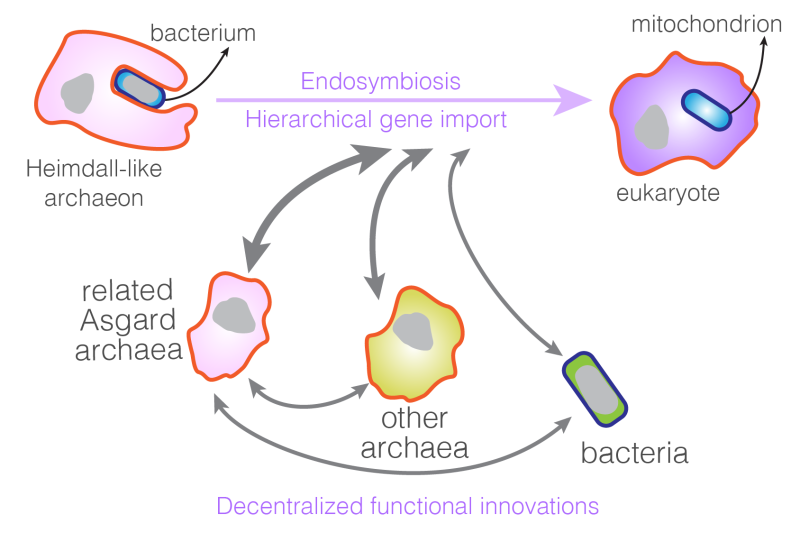
Figure 1: A simple depiction of a model describing the horizontal gene transfer events (grey arrows) that led to the emergence of eukaryotic complexity.
Combining a year-long anaerobic cultivation and Oxford nanopore long-read sequencing, Wu and colleagues for the first time recovered the circular genomes of two species named Candidatus Heimdallarchaeum endolithica and Heimdallarchaeum aukensis, as well as several other near-complete genomes from related Asgard lineages. These genomes not only provided solid evidence that confirmed the close relationship between Heimdallarchaeota and the eukaryotes, but also captured repetitive sequences that had been otherwise lost or misassembled. They include CRISPR/Cas operons, which contain CRISPR spacers that enabled the discovery of mobile elements targeting Asgard archaea for the first time. For example, a new class of mobile elements - Aloposons, named after the twin giants in Greek mythology, was found to encode two giant proteins containing around 5000 amino acids. Asgard archaea mobile elements inherited diverse nucleic acid-processing enzymes from bacteria and bacteriophages, indicating a complex evolutionary entanglement between Asgard archaea and bacteria.
It has been found that the eukaryotic genomes garnered their gene content from both bacterial and archaeal domains, and curiously, the fraction of bacteria-derived genes increases with genome size. This study shows a quantitative agreement between Asgard archaea and eukaryotes both in terms of gene numbers and functional division, as well as a striking difference between the scaling properties of the ‘bacterial’ faction and ‘archaeal’ fraction in Asgard archaea genomes. The observed parallelism suggests that understanding how Asgard archaea (or archaea in general) regulate their genome size and the gene content may provide a clue for the early stages of gene acquisition that established the eukaryotic genomes. Based on further quantitative analyses on the genes shared between Asgard archaea and the eukaryotes, Wu and colleagues put forth a conceptual framework called the Heimdall nucleation - Decentralized innovation - Hierarchical import (HDH) model to account for the horizontal gene transfer events that led to the emergence of eukaryotes.
|
HFSP award information Long-Term Fellowship (LT000468/2017-L): Energy budgeting of microbial players in global carbon and nitrogen cycles Fellow: Fabai Wu |


































