Proteins carry out most of the chemical tasks required for life. In many cases, the biological function of a protein is linked to a well-defined three-dimensional structure or to an ensemble of closely related structures. In modern organisms, the folding to the functional structures of the polypeptide chains emerging from the ribosome is guided and assisted by the coordinated action of a diversity of molecules called chaperones. However, molecular chaperones are themselves a complex product of evolution and, since evolution has no foresight, molecular folding-assistance should not have preceded protein self-folding. This logic suggests that the most ancient proteins could plausibly fold efficiently by themselves.
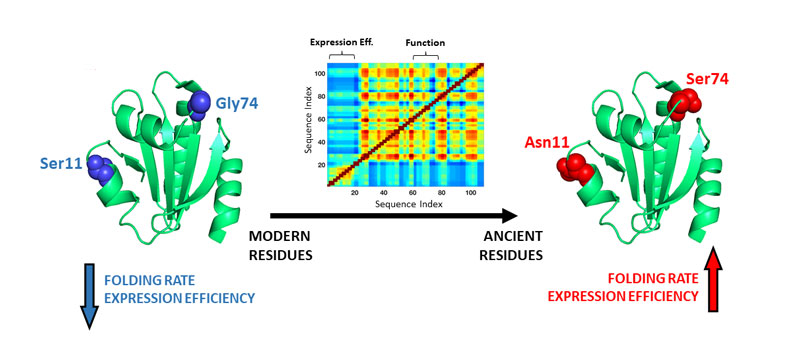
Figure: Enhancing proteins through computationally-guided, back-to-ancestor engineering.
Obviously, ancient proteins belong to extinct organisms that no longer exist on Earth. Yet, the exponentially-growing sequence databases of the post-genomic era, together with advances in bioinformatics and phylogenetic analyses, allow the derivation of plausible approximations to the sequences of proteins that existed millions and even billions of years ago. Standard molecular biology methodologies can then be used to prepare in the lab the proteins encoded by the reconstructed sequences (the “resurrected” ancestral proteins). It thus becomes possible to experimentally study how the properties of ancestral proteins differ from those of their modern counterparts and to explore the evolutionary and biotechnological implications of such differences. Combining the expertise of Eric Gaucher (USA) and Jose M. Sanchez-Ruiz (Spain) through an HFSP grant enabled these researchers and their colleagues to explore the unique combination of evolutionary engineering.
Obligate symbionts, i.e., organisms that depend entirely on other organisms for survival, typically display high evolutionary rates. Comparison of their proteins with the corresponding protein ancestors should highlight changes during evolution at both the sequence and biomolecular property levels. Thioredoxins function as general redox catalysts in all known cells. We have found an enormous difference between the folding behaviour of ancient thioredoxins from billion-year-old phylogenetic nodes compared to a modern thioredoxin from the luminous bacterium Candidatus Photodesmus katoptron, the obligate symbiont responsible for the light emitted by the sub-ocular organs of flashlight fish. Folding of the symbiont thioredoxin is severely impaired outside its original host, as shown by a very slow in vitro (i.e., in the test tube) folding and by an inefficient heterologous expression within E. coli that leads mostly to insoluble protein. By contrast, the resurrected Precambrian thioredoxins fold fast in vitro and efficiently within the heterologous host. Experimental mutational analyses guided by computational folding simulations allowed us to identify the sequence changes responsible for the folding differences and to engineer efficient folding in the symbiont protein on the basis of only a few back-to-the-ancestor mutations.
Our results have both evolutionary and biotechnological implications. From an evolutionary point of view, they highlight co-evolution of proteins with the folding-assistance machinery of their natural hosts, while supporting that very ancient protein folding was indeed unassisted to a substantial extent. In addition, our results demonstrate an ancestral-reconstruction approach to rescue inefficient heterologous expression, a major biotechnological bottleneck. Since the approach involves limited sequence engineering, it may be particularly useful in sequence-screening metagenomics efforts targeting recent adaptations. The efficient preparation of pollutant-degrading enzymes from uncultured organisms growing at contaminated sites is a potential application of particular interest.
|
HFSP award information Research Grant - Program (RGP0041/2017): Generating and understanding de novo enzyme functionalities using ancestral proteins as scaffolds Principal investigator: Jose M. Sanchez-Ruiz, University of Granada, Spain |


































