Phenomena such as symmetry breaking and spatiotemporal patterning of different cell types have historically been explained from a gene-centric perspective. In this view, specific genes, organized into networks with defined topologies, sense internal and external inputs to drive the complexity characteristic of multicellular organisms. However, the role of cell density in these processes has received relatively little attention.
Theoretically, cell density can influence several aspects of cellular behavior: it can alter cell shape, affecting the strength of intercellular communication; it can modify cell movement, changing the number of neighboring cells with which a cell can interact; and it can impact the metabolic state of cells, as competition for limited nutrients in crowded populations can slow down their metabolism, among other effects.
In a study published in Nature Communications, the HFSP Fellowship Awardee Marco Santorelli and team investigated how cell density affects the patterning of a synthetic gene regulatory network based on a synthetic Notch receptor. Specifically, they generated two distinct cell lines: a "sender" cell line stably expressing a synthetic Notch ligand and a "transceiver" cell line expressing a synthetic Notch receptor that, upon activation, drives the expression of its own ligand. In theory, such a network, lacking a genetically encoded negative feedback mechanism, should enable unlimited propagation of the synthetic Notch signal, leading to activation of the entire cell population.
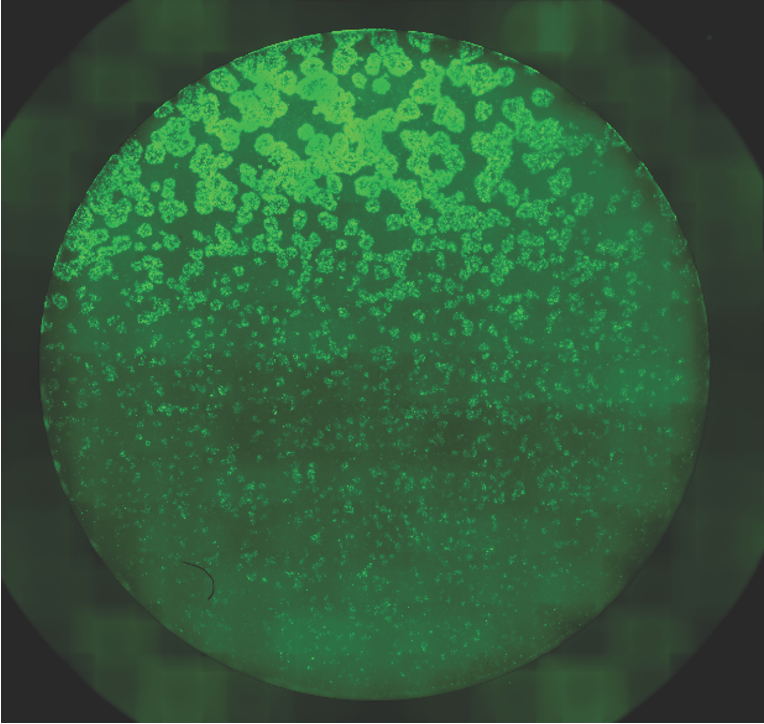
Surprisingly, the researchers found that synthetic Notch activation was highly dependent on cell density, which acted as a non-genetically encoded negative feedback mechanism. As cell density naturally increased during the experiment (due to cell replication), signal propagation became self-limiting, resulting in the formation of placode-like spots. The size of these spots could be precisely controlled by modulating the initial cell density or by adjusting cellular growth rates through drug treatments.
Another significant finding was that seeding cells along a density gradient produced millimeter-scale activation patterns. These consisted of a polarized layer of cells, with a "head" of non-activated cells and a "tail" where the synthetic Notch signal was active. Furthermore, by dynamically modulating the density gradient in both space and time, the group led by the HFSP Fellow was able to generate a kinematic wave of activation.
Altogether, these findings present a novel approach for studying and controlling patterning circuits by manipulating cell growth and density.


































