The genetic code is degenerate. Most amino acids are encoded by multiple, so-called synonymous codons. Synonymous codons were initially presumed to have entirely equivalent functions. However, synonymous codon usage is biased, as abundant and rare codons are distributed non-randomly in whole genomes and along the open reading frames of genes. Thus, codon choice might have functional implications beyond amino acid coding. In fact, alterations in codon usage patterns due to synonymous mutations can lead to disease. Synonymous codons may modulate protein folding by tuning kinetics of translation. However, the exact molecular mechanism connecting codon usage to protein homeostasis is not known.
To understand the link between synonymous codon usage, translation, and protein folding, we utilized mammalian eye lens gamma-B crystallin as a model protein and investigated its expression in vitro and in vivo in Escherichia coli. Due to differences in codon usage biases between the native gamma-B crystallin host, Bos taurus and E. coli, the codon distribution in the native gamma-B crystallin mRNA sequence is non-optimal for translation in E. coli. We therefore designed two variants of the mRNA coding for gamma-B crystallin, one with the codon usage that would be optimal for protein translation in E. coli (with an mRNA codon distribution similar to that found in B. taurus, which was expected to result in more natural translation kinetics) and the other with unaltered codon composition un-optimal for translation in E. coli. Expression of the optimal variant yielded more soluble and protease-resistant protein than that of the variant with non-optimal codon distribution along the gamma-B crystallin mRNA. Importantly, both variants had identical amino acid sequences. We then probed the structure of purified, mature proteins by 2D NMR and detected considerable structural heterogeneity of the synonymous gamma-B crystallin variants arising from different oxidation states of a cysteine pair in the N-terminal domain of the protein. We concluded that the ‘silent’ polymorphisms altered the distribution of populations within the landscape of accessible protein conformations. Protein expressed from the optimal variant was found to be more similar to its natural analog isolated from bovine eye lenses.
Click on image to enlarge
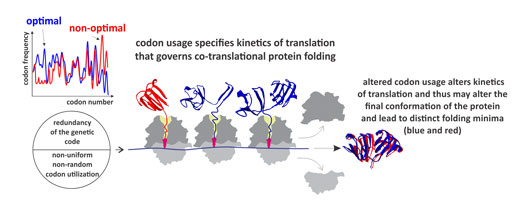
Figure: Synonymous codon usage alters kinetics of protein translation and directs co-translational folding towards different protein conformations. The appearance of distinct folding intermediates is linked to the overall and local rate of translation. Altered codon usage results in modified translation kinetics and folding events on the ribosome (blue and red), which may lead to altered final protein conformations.
We next asked whether codon usage affects translation. To address this question, we monitored real-time translation kinetics in a fully-reconstituted rapid in vitro translation system. The mRNA variant with non-optimal codon distribution was translated more slowly than the variant with the near-native codon usage. Slower translation may result in a delayed onset of protein folding, because it takes more time for the protein to emerge from the ribosome. However, this does not explain the dramatic effect on the protein conformation and stability. To monitor co-translation folding and understand the nature of conformational changes, we developed an in vitro Förster resonance energy transfer (FRET) approach to monitor distance changes within the emerging nascent chain during on-going translation. We show that the presence of rare codons changes the rate of folding of the N-terminal protein domain after it emerged from the exit tunnel. The speed of translation may define the window of opportunity for the protein parts to fold locally, particularly at the critical points where folding is far from equilibrium. These observations provide a strong support to the hypothesis that synonymous codons usage serves as a secondary code for protein folding in the cell.
This work deepens our understanding of protein folding, one of the most fundamental mechanisms in the cell. It may also help to explain how genetic diseases linked to silent mutations develop, which may potentially allow physicians to create more specific, personalized treatments based on an individual’s genetic information. Finally, this work may give a tool to upscale the production of functional active recombinant proteins for medical and biotechnological purposes.
Reference
Synonymous codons direct cotranslational folding toward different protein conformations. Florian Buhr1†, Sujata Jha2†, Michael Thommen3†, Joerg Mittelstaet3, Felicitas Kutz1, Harald Schwalbe1*, Marina V. Rodnina3* and Anton A. Komar2,4,5* Molecular Cell, 61 (3), 341–351, 2016. †Co-first authors; *Co-corresponding authors.