The discovery of the molecular mechanism of the circadian clock was recognised by the Nobel Prize in Physiology or Medicine in 2017. From what we know, it is not a single centralized clock that is responsible for our circadian cycles. Instead, it operates in a hierarchical network with a “master pacemaker” and a “slave oscillator”. The master pacemaker receives various input signals from the environment such as light. It then drives the slave oscillator that regulates various outputs such as sleep, feeding, and metabolism. Despite the different roles of the pacemaker neurons, they are known to share common molecular mechanisms that are well conserved in all lifeforms. For example, interlocked systems of multiple transcriptional-translational feedback loops (TTFLs) composed of core clock proteins have been extensively studied in fruit flies.
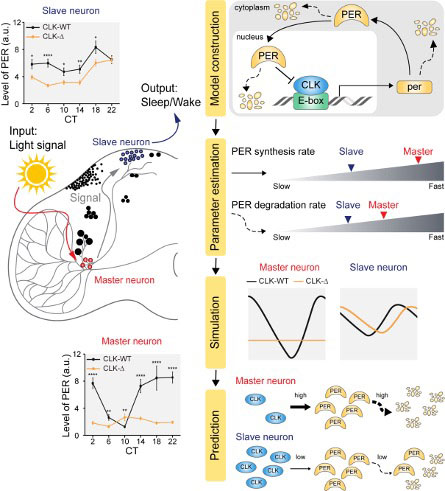
Fig. 1: (left) The circadian clock operates as a network where the master pacemaker and slave oscillator are organized in a hierarchical manner. Although they generate rhythms via an identical transcriptional-translational feedback loop (TTFL) of PER proteins, the generated PER rhythms are different between them. (right) By using a mathematical model describing the TTFL of the Drosophila circadian clock (top), we identified the differences between master and slave clock neurons (bottom).
However, there is still much that we need to learn about our own biological clock. The hierarchically-organized nature of master and slave clock neurons leads to a prevailing belief that they share an identical molecular clockwork. At the same time, the different roles they serve in regulating bodily rhythms also raise the question of whether they might function under different molecular clockworks.
Led by Profs Jae Kyoung Kim (IBS & KAIST) and Eun Young Kim (Ajou University), researchers used a combination of mathematical and experimental approaches using fruit flies to answer this question. The team found that the master clock and the slave clock operate via different molecular mechanisms.
In both master and slave neurons of fruit flies, a circadian rhythm-related protein called PER is produced and degraded at different rates depending on the time of the day. Previously, the team found that the master clock neuron (sLNvs) and the slave clock neuron (DN1ps) have different profiles of PER in wild-type and Clk-Δ mutant Drosophila. This hinted that there might be a potential difference in molecular clockworks between the master and slave clock neurons.
However, due to the complexity of the molecular clockwork, it was challenging to identify the source of such differences. Thus, the team developed a mathematical model describing the molecular clockworks of the master and slave clocks. Then, all possible molecular differences between the master and slave clock neurons were systematically investigated by using computer simulations. The model predicted that PER is more efficiently produced and then rapidly degraded in the master clock compared to the slave clock neurons. This prediction was then confirmed by follow-up experiments using animals.
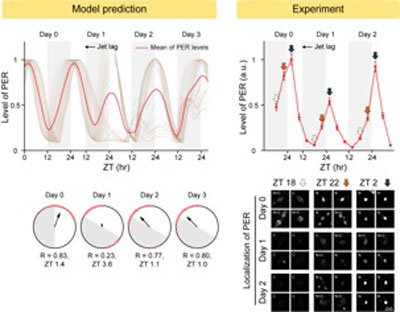
Fig. 2: The combination of mathematical model simulations and experiments revealed how the master clock neurons can obtain both robustness (i.e., high amplitude and period robustness) and plasticity (i.e., fast and wide range of entrainment). (left) The model predicts that when jet lag occurs, the master clock neurons dispersed the phase of rhythms, reducing the amplitude of rhythms. This leads to the rapid adaption to the new time zone. (right) This model prediction was confirmed by the follow-up experiments.
Then, why do the master clock neurons have such different molecular properties from the slave clock neurons? To answer this question, the research team once again used the combination of mathematical model simulation and experiments. It was found that the faster rate of synthesis of PER in the master clock neurons allows them to generate synchronized rhythms with a high level of amplitude. Generation of such a strong rhythm with high amplitude is critical to delivering clear signals to slave clock neurons.
However, such strong rhythms would typically be unfavorable when it comes to adapting to environmental changes. These include natural causes such as different daylight hours across summer and winter seasons, up to more extreme artificial cases such as jet lag that occurs after international travel. Thanks to the distinct property of the master clock neurons, it is able to undergo phase dispersion when the standard light-dark cycle is disrupted, drastically reducing the level of PER. The master clock neurons can then easily adapt to the new diurnal cycle. Our master pacemaker’s plasticity explains how we can quickly adjust to the new time zones after international flights after just a brief period of jet lag.
It is hoped that the findings of this study can have future clinical implications when it comes to treating various disorders that affect our circadian rhythm. Jae Kyoung Kim notes, “When the circadian clock loses its robustness and flexibility, the circadian rhythm's sleep disorders can occur. As this study identifies the molecular mechanism that generates robustness and flexibility of the circadian clock, it can facilitate the identification of the cause of and treatment strategy for the circadian rhythm sleep disorders.”
|
HFSP award information Research Grant - Early Career (RGY0063/2017): The molecular circadian clock as a causal mediator of sleep-regulated neurophysiology and cognition Principal investigator: Robbert Havekes, Groningen Institute for Evolutionary Life Sciences (GELIFES), University of Groningen, Netherlands |


































