Abundance of genes in a population is captured by the frequencies of alleles at the corresponding genomic positions. Selection, mutation and randomness due to finite population size affect the dynamics of allele frequencies and consequently of quantitative traits. While the macroscopic dynamics of quantitative traits can be observed, the microscopic allele frequencies are typically unobserved. The fundamental question is then whether we can predict the dynamics of macroscopic variables without information on the microscopic dynamics.
Evolution of quantitative traits can be described by the maximum entropy formalism used in many other complex systems, for example bird flocking or fish swarming. However, our approach relies, in addition, on a well-defined microscopic model of allele frequency dynamics, which is known for evolutionary problems but not for many other complex systems. This microscopic model, the so-called Fokker-Planck or diffusion equation, in principle contains all the information we care about, but is intractable to solve directly. We show that the maximum entropy principle can be used to construct a very good and computationally tractable approximation to the Fokker-Planck equation, by only focusing on the dynamics of macroscopic observables (quantities, which are in principle measurable) that we care about. By choosing the correct set of observables (trait mean, mean heterozygosity) our approach recovers the correct stationary distribution. In our approximation, however, this stationary state is allowed to change in time in response to the external forces (mutation and selection) by tuning a small number of relevant parameters; we call them “effective evolutionary forces”. These parameters are made to change in time such that the dynamics of observables agrees with the solution of the Fokker Planck equation. The resulting dynamics of the tuning parameters are low dimensional and interestingly, do not depend on the microscopic details of the full model.
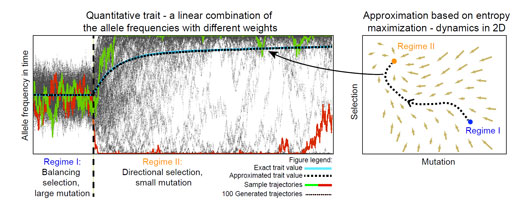
Figure: Quantitative traits depend on frequencies of alleles in the individual genomes in the population. These change in time due to changes in evolutionary forces, leading to the dynamics of the trait. The dynamics in this example are caused by a rapid change in evolutionary parameters at the beginning of regime II when both the selection and the mutation are perturbed at the same time. Our approximation dramatically simplifies the complex dynamics while retaining excellent accuracy.
Our work shows that even when the evolutionary forces, which drive the evolution of quantitative trait, change rapidly, our simple approximation captures the macroscopic quantities extremely well. This is true even when the trait depends on many genes and their effect on the trait greatly differs. Moreover, our method, unlike the previously used methods, works in the realistic regime of small mutation. Despite its remarkable accuracy and its potential to extend to a broad class of non-stationary problems across the spectrum of biology and physics, approximations of this type have not been noticed either by physicists or by biologists, and remain to be explored further.
Reference
A general approximation for the dynamics of quantitative traits, Boďová, K., Tkačik, G., Barton, N., Genetics, 2016 202: 1-26.