How the vast diversity of neurons in the brain are generated is a central question in neurobiology. The mechanism by which neural stem cells generate different neurons with time is called temporal patterning. This process is conserved in mammals, but it is better understood in Drosophila, where a series of tTFs sequentially expressed in neural progenitors as they age generate different identity windows in which different neurons are generated. Earlier screens for tTFs in the medulla (one of the optic lobe neuropils, which processes visual information) were limited by antibody availability, and the six previously identified tTFs could not account for the generation of the complete neuronal diversity in this brain region.
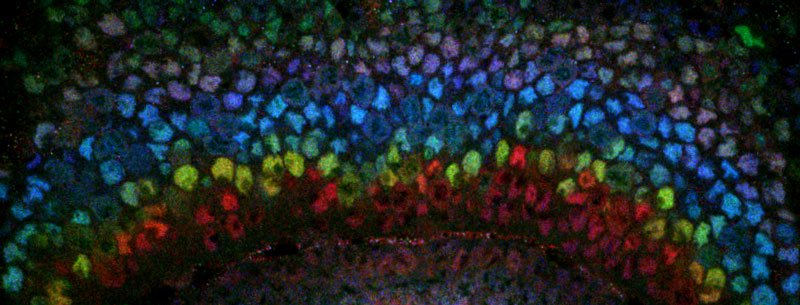
Figure: Confocal microscopy image of the developing fly optic lobe in which neural stem cells express different temporal transcription factors (shown in different colors) as they age to generate different neurons with time. Credits: Isabel Holguera, Claude Desplan laboratory, New York University.
In this work, we used state of the art single-cell RNA-sequencing (scRNA-seq) of the developing Drosophila optic lobe to interrogate the complete set of tTFs that pattern medulla neural progenitors in a comprehensive and unbiased way. We identified seven previously undescribed tTFs and validated their expression pattern relative to the previously known tTFs in vivo. Moreover, we determined the genetic interactions between these and the previously described tTFs that allow the temporal cascade to progress, validating the notion that a given tTF activates the expression of the next tTF in the cascade and represses the expression of the previous one, and uncovering more complex regulations.
We characterized the role of different tTFs in the generation of neural diversity by analyzing how the generation of specific neuronal progeny is affected in their absence. Furthermore, we made use of TFs expressed in medulla neurons in a temporal manner informed from the scRNA-seq dataset to assign the birth order and temporal window of origin of all the medulla neurons. Importantly, the comprehensive list of tTFs described in this work, together with spatial patterning of the neuroepithelium that give rise to medulla neural stem cells and differential Notch signaling in the neuronal progeny, can explain the generation of all the neuronal diversity produced in this brain region.
Finally, we characterized the first steps of neuronal differentiation in fly optic lobe neurons and found that this process is similar in human cortical neurons, with similar groups of genes being expressed during this process.
These results open new avenues to understanding why neurons are generated in a stereotyped order and could have implications for regenerative medicine in the future: knowing the molecular players acting on stem cells to generate neural diversity will allow us to activate neural stem cells in vivo to replace missing neurons or repeat these developmental processes in vitro for cell replacement therapy.
In the Media
Harvard Medical School News Release
National Eye Institute Research News
Genetic Engineering & Biotechnology News
|
HFSP award information Long-Term Fellowship (LT000757/2017-L): Orderly assembly of neurons in the Drosophila motion vision circuits Fellow: Isabel María Holguera López |


































