Many bacteria primarily exist in nature as multicellular communities, so called biofilms, which are held together by a self-produced extracellular matrix. Although biofilms are desirable in waste-water treatment applications, biofilms primarily cause undesirable effects such as chronic infections or clogging of industrial flow systems. Cells in biofilms display many behavioural differences from planktonic cells, such as an increased tolerance to antibiotics, an altered transcriptome, and spatially heterogeneous metabolic activity.
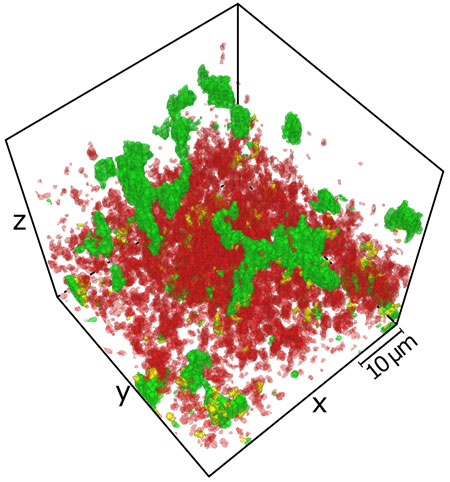
Figure: Individual cells in E. coli biofilms display mutually exclusive expression of curli amyloid fibers (green) and flagella (red).
Biofilm formation is a highly regulated and dynamic process that includes the transition from the motile planktonic to sessile biofilm lifestyle. Growth of planktonic cells into biofilm communities critically depends on the production of extracellular matrix. The exact composition of the extracellular matrix varies from species to species, but for E. coli the matrix primarily includes curli amyloid fibers, flagella, cellulose, and other polysaccharides.
Cellular differentiation within a biofilm is a commonly accepted concept, but it remains largely unclear when, where and how exactly such differentiation arises. In this study we quantitatively analyzed spatio-temporal changes in expression of several key groups of E. coli genes, including matrix-production genes, during the formation of biofilms. The expression of genomic fluorescent reporters was studied at the single-cell level, using both flow cytometry and image analysis of the biofilm structures. We showed that while expression of the curli and flagellar genes is confined to distinct subpopulations of cells, other gene classes are expressed more uniformly across the subpopulations. Furthermore, using molecular timers we demonstrated that growth rates of cells vary substantially between different regions of the biofilm. Although the physiological importance of phenotypic heterogeneity in E. coli biofilms, and its regulation by the environmental and growth conditions, needs further investigation, the observed diversification appears to be spontaneous rather than being driven by nutrient or other gradients.
This study demonstrates the existence of several specialized cell subpopulations in E. coli biofilms, which are distinctly different in their gene expression profiles and presumably in their physiological state. These subpopulations critically impact on the three-dimensional community architecture.
Reference
Diversification of Gene Expression during Formation of Static Submerged Biofilms by Escherichia coli. Besharova O, Suchanek VM, Hartmann R, Drescher K, Sourjik V. Front. Microbiol. 7:1568 (2016). doi: 10.3389/fmicb.2016.01568


































