To truly understand how development proceeds and how cells form tissues, it is very important to observe cell and tissue biology when it happens and where it happens in the unperturbed embryo. However, it becomes more and more clear that confocal systems, that have been widely used for such studies come with certain limitations, one of the major ones being the induction of phototoxicity. To this end, the development of light sheet microscopy was an important step to reduce this problem and allow for long, fast and gentle imaging of many tissues in many different organisms. One of the organisms best suited to light sheet imaging is the translucent zebrafish embryo. Organ development is often conserved between the zebrafish and higher vertebrates, making it an ideal model to understand cell biology and tissue mechanics of these processes that can later be transferred to other less accessible systems or ex vivo models. However, one obstacle that needs to be overcome to generate meaningful conclusions from the data is to manage the immense data amounts that result from light sheet experiments. Our protocol deals on one hand with the details of setting up an imaging experiment and on the other, makes suggestions how to analyze the resulting data.
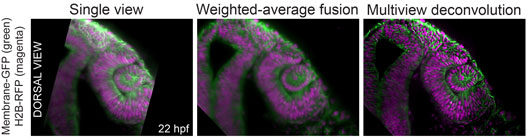
Figure: A quality comparison between single view images and data processed by two different multi-view fusion algorithms. The image shows a developing zebrafish eye and neural tube.
Our open access video protocol (Icha, Schmied et al., 2016) establishes a versatile light sheet microscopy experimental pipeline. This pipeline is based on a commercial microscope and an open source software solution for data processing. Specifically, we used the Lightsheet Z.1 microscope from Zeiss and the Multiview reconstruction application in Fiji to process the data. The implications of this approach are demonstrated by imaging several stages of retinal development in zebrafish spanning from optic cup formation to neuronal translocation. However, this pipeline can be used for long-term time-lapse imaging of a plethora of morphogenetic events in different model organisms. Overall, the protocol covers the essential steps of a light sheet microscopy experiment from mounting the sample to processing the data. It needs to be noted though that the most complicated step in light sheet microscopy is not acquiring the images but the subsequent read out of the information in the data, especially, when multiple views are imaged and need to be fused. The solution provided in the protocol is to embed fluorescent beads as fiduciary markers around the sample to register the different views onto each other, which allows the reconstruction of the entire imaged volume of the sample. For fast and interactive visualization of the often very large datasets we are using the BigDataViewer in Fiji.
Overall, this protocol shows the cross disciplinary nature that is important when acquiring light sheet data as developmental biology needs computer scientists. With this hands-on manual it should also be easy for first time users to get over initial reserve and start imaging in order to ask the questions in developmental biology that were much more difficult to ask previously.
Reference
Using Light Sheet Fluorescence Microscopy to Image Zebrafish Eye Development. Icha, J., Schmied, C., Sidhaye, J., Tomancak, P., Preibisch, S., Norden, C. (2016) J Vis Exp. 2016 Apr 10;(110). doi: 10.3791/53966.


































