The roundworm C. elegans is one of the most widely used and most versatile model organisms, and it is especially useful for revealing principles of animal development. It is optically transparent, exhibits an invariant cell lineage, and can be functionally probed with modern genetics. Studies of embryonic and larval C. elegans development have transformed and deepened our understanding of developmental mechanisms. However, long-term high-resolution in vivo imaging of these processes has been possible only in embryos.
Why is time lapse microscopy of C. elegans post-embryonic development so difficult? First, newly hatched larvae are only 15μm wide and 250μm long; adult animals are much larger, typically 40μm wide and 1,000 μm long. Second, larvae require food for growth to progress through normal development (~50hrs), and locomotion to shed and replace their cuticle at each larval molt. Automated methods for imaging must, therefore, contend with animal growth, and must allow sufficient locomotion for molting.
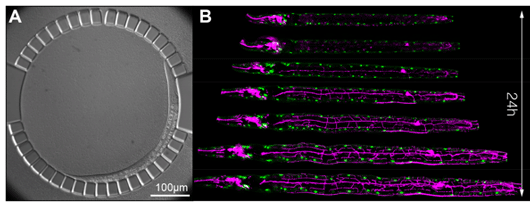
Figure: High-resolution live imaging of C. elegans larvae in a microfluidic chamber.
(A) Differential interference contrast (DIC) image of an immobilized larva in the microfluidic chamber. For its entire post-embryonic development, the animal is confined in a circular array of posts with 4.88-μm spacing. Entrance to the chamber (top) is 10 μm wide. The animal moves freely in the chamber and is periodically immobilized to allow for high-resolution z-stack acquisition. (B) Computationally aligned time lapse fluorescence micrographs of a C. elegans larva, expressing fluorescent reporters labeling all body wall muscle nuclei (green) as well as a nociceptive neuron called PVD (magenta).
To overcome these challenges, Keil et al. developed a simple microfluidic device, where up to ten animals can be grown in micro-chambers and can be periodically immobilized to allow for the acquisition of high-resolution z-stacks as frequently as every 4 minutes (Figure A). The authors demonstrate the versatility of their technique by collecting large numbers of cell lineages and quantifying temporal dynamics of reporter gene expression during a variety of developmental events that hitherto had been inaccessible to time-lapse microscopy. For instance, the authors obtain cell-cycle statistics for the development of the worm vulva - a paradigm for organogenesis – in over one hundred animals. Due to the large number of cell lineages, Keil et al. are able to identify statistically significant trends in cell-cycle onsets and lengths and reveal rare deviations from the generally invariant cell lineage in the worm. Finally, the authors develop computational methods to automatically align consecutive z-stacks. Combining long-term imaging with computational alignment is then able to provide spectacular views of post-embryonic animal development such as the outgrowth of complex networks of dendritic branches in C. elegans touch neurons (Figure B).
This new method will enable developmental studies in C. elegans larvae - freely moving and growing animals - with unprecedented temporal resolution and control. Such studies are likely to reveal unexplored phenomena and to contribute to our understanding of general mechanisms governing metazoan development at the cellular level.
Reference
Long-Term High-Resolution Imaging of Developing C. elegans Larvae with Microfluidics. Keil, W., Kutscher, L. M., Shaham, S., & Siggia, E. D. (2017). Developmental Cell, (40), 1–13.


































