Structural transitions of chromatin are central to eukaryotic gene regulation. A key mediator of such transitions is chromatin remodeling, a process in which different classes of remodeling enzymes reposition nucleosomes using the energy of ATP hydrolysis. Little is known about the conformational state of the histone octamer core of a nucleosome during remodeling reactions due to lack of structural data and limitations of biochemical techniques.
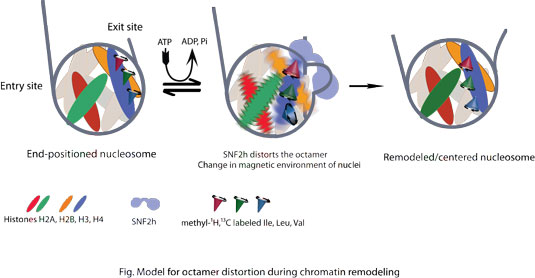
In the present study, we have leveraged the power of methyl- transverse relaxation optimized spectroscopy (TROSY) NMR to probe the effect of a remodeling enzyme, SNF2h, on the dynamics of internal amino acid residues that are normally buried within a nucleosome. To do this, terminal methyl groups of Isoleucine, Leucine and Valine (ILV) residues in individual histones have been magnetically (13C, 1H) labeled using metabolic precursors in an otherwise completely deuterated background. Two-dimensional NMR spectrum of nucleosomes containing perdeuterated histone octamer with methyl-ILV labels on histone H4 has been obtained both alone and in the presence of an activated form of SNF2h. The spectrum of the 450 kD nucleosome-SNF2h complex in the presence of a non-hydrolysable transition-state ATP mimic, ADP.BeFx, suggests that several ILV residues buried within the core are differentially broadened. The broadening is indicative of dynamics on ms-µs time scale due to a change in the magnetic environment of the corresponding ILV residues. These results indicate that the octamer is distorted by SNF2h. The NMR spectra of nucleosomes methyl labeled at Isoleucines of histone H2A suggest that SNF2h produces similar dynamics in H2A. Because H2A makes minimal contacts with the H3-H4 interface, it appears that the observed distortion encompasses a large region of the nucleosome.
Using site-specific disulfide bridges in two distinct locations in the H3-H4 interface, we show that interfering with octamer deformability inhibits nucleosome sliding by SNF2h. In contrast, a different class of remodeler, INO80, can slide these cross-linked nucleosomes. RSC, a member of the SWI/SNF family, is seen to slide the cross-linked nucleosomes, and additionally, also shows strikingly increased octamer eviction. Thus, it appears that different classes of remodeling enzymes can exploit octamer deformability in different ways.
By showing that the histone octamer is deformable these results have opened up an exciting new way to understand different transactions of chromatin. Importantly, the results have brought us a step closer to understanding the molecular determinants of the intricate process of euchromatin-heterochromatin transition. It is also possible that the plasticity of histone octamer plays key roles in other fundamental processes such as transcription, recombination and repair.
Reference
Distortion of histone octamer core promotes nucleosome mobilization by a chromatin remodeler. Sinha K.K., Gross, J.G. & Narlikar G.J. Science, 355, eaaa3761 (2017).


































