Sexual reproduction requires the formation of haploid gametes from diploid precursor cells by a specialized cell division process called meiosis. Errors in this process are a major cause of infertility, and can also cause developmental disorders such as Down syndrome. To ensure proper segregation of homologous chromosomes during meiosis I, chromosomes must first pair during meiotic prophase. Pairing is then stabilized by the formation of the synaptonemal complex and crossover formation, which introduces physical linkages through homologous recombination. All of these processes rely on the dramatic re-organization of meiotic chromosomes into linear arrays of loops anchored to a central axis, which occurs at the beginning of the meiotic cell cycle program. Chromosome axes, which contain both cohesins and additional meiosis-specific proteins, play essential roles in homolog pairing, synapsis, and recombination. Although the proteins that comprise the axis have been identified in several organisms, their organization within this structure remains unknown. To better understand axis function, we sought to define its molecular organization in C. elegans.
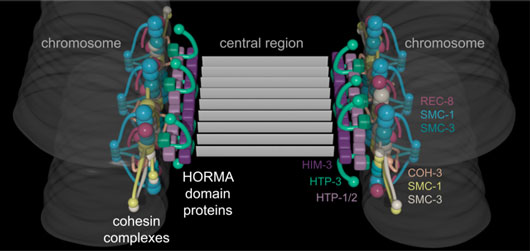
Figure: A model of chromosome axes based on three-dimensional super-resolution imaging. Colors correspond to individual proteins as indicated in the figure.
The size scale of the axis makes it inaccessible to conventional light microscopy, but super-resolution techniques such as STORM (STochastic Optical Reconstruction Microscopy) are able to localize proteins to a resolution of tens of nanometers, or sometimes better. As chromosomes find their partners, each axis becomes aligned with that of the homologous chromosome through the formation of the synaptonemal complex, a protein scaffold that assembles between them and holds them at a well-defined distance of ~150 nm. This regular spacing and symmetry make it possible to average over many images to define the positions of individual proteins at very high (sub-10-nm) precision. By combining our results with known structures and interactions of axial proteins, we were able to develop a three-dimensional model of chromosome axes (see Figure).
We found that cohesin complexes and axis-associated meiosis-specific “HORMA” domain proteins are located within distinct substructures of the axis. Cohesins lie farther from the midline between homologous chromosomes, while the HORMA domain proteins appear to bridge the gap between cohesins and the synaptonemal complex. Thus, our data reveal how these meiosis-specific proteins establish a platform for the formation of the synaptonemal complex. Moreover, we demonstrated that two different types of cohesin complexes that localize to the axis have distinct arrangements within this structure (see Figure): REC-8 containing cohesin complexes protrude above and below the central plane of the axis/synaptonemal complex while COH-3 containing complexes are in the center. We speculate that this organization of cohesin complexes creates a physical barrier to inter-sister recombination, thereby promoting inter-homolog recombination. This homolog bias is thought to promote crossover formation, which is absolutely required to accurately align and segregate homologous chromosomes in meiosis I and generates new combinations of traits for future generations.
Reference
Superresolution microscopy reveals the three-dimensional organization of meiotic chromosome axes in intact Caenorhabditis elegans tissue. Köhler, S., Wojcik, M., Xu, K. & Dernburg, A. F. Proc. Natl. Acad. Sci. 114, E4734–E4743 (2017).


































