If the DNA in a single human were stretched end-to-end it would stretch over 2 meters, but the genome still manages to fit into a tiny volume of the nucleus in the cell. To achieve this, DNA is heavily compacted around chromatin proteins. The basic unit that wraps and compacts the DNA is an octamer of histone proteins called the nucleosome. The nucleosome doesn’t just compact the DNA, but also helps to decide how the information in the DNA is used. For instance, if the DNA becomes broken or damaged, nucleosomes near the damaged site become heavily post-translationally modified, highlighting the area to the specialized repair machines of the cell. How modified nucleosomes are read on DNA has been an elusive unanswered question to date.
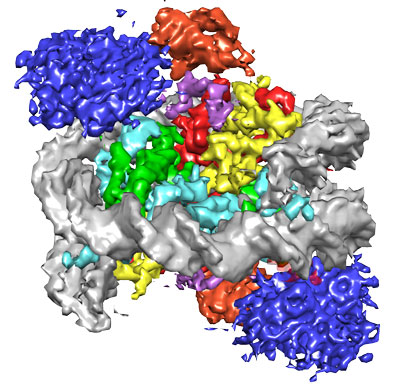
Figure: Representation of NCP-ubme-53BP1 structure at 4.5 Å resolution. Histone proteins are coloured green, cyan, red and yellow and DNA grey. Blue and purple 53BP1 is sandwiched next to constrained ubiquitin, coloured orange.
Recent work by Wilson and colleagues aimed to look at how the chemical tags that occur around broken DNA are read by a key DNA repair protein, called 53BP1. 53BP1 is recruited to sites of DNA breaks, but does not bind to the damaged DNA directly. Instead it recognises modified nucleosomes near the site of damage, by an unknown molecular mechanism. The study investigated how 53BP1 can be so specific to the tagged nucleosomes at DNA breaks, by making artificially modified nucleosomes and mixing them with a fragment of 53BP1. They found that they interacted directly and went further to look at the molecular details of this interaction by electron cryomicroscopy.
The structure of 53BP1 bound to a modified nucleosome revealed that 53BP1 forms direct contacts with histone tail methylation and ubiquitylation modifications, as well as the nucleosome surface itself. While methylation binding is limited to just the histone tail, 53BP1 was found to specifically sandwich itself between the nucleosome surface and ubiquitin, fixing ubiquitin into a constrained conformation. This recognition garners both high affinity and high selectivity to chromatin around the sites of DNA DSBs.
This 3D model offered novel insights into how 53BP1 binds to multiple elements on both the ubiquitin and the nucleosome itself to determine how 53BP1 responds to DNA breaks. This wasn’t just an academic exercise; how different types and combinations of post-translational modifications are recognised is key to understanding the control of almost all chromatin processes in the cell and has been poorly understood up until now.
Reference
The structural basis of modified nucleosome recognition by 53BP1. Wilson MD*, Benlekbir S*, Fradet-Turcotte A, Sherker A, Julien JP, McEwan A, Noordermeer SM, Sicheri F, Rubinstein J, Durocher D. Nature, 536 Aug 4 pg 100-103.


































