The discovery that new genes can emerge from previously non-functional DNA is transforming our understanding of how life innovates. In the current study, HFSP Fellowship Awardee Idan Frumkin and his host supervisor, HFSP Research Grant Awardee Michael T. Laub, recreated the very beginning of gene evolution in the laboratory by testing more than one hundred million completely random genetic sequences. Remarkably, thousands of these “proto genes” were immediately useful to bacteria when challenged with a deadly virus. This shows that the raw material for evolutionary innovation may be far more abundant and accessible than previously thought, and that even unevolved sequences can produce meaningful biological functions.
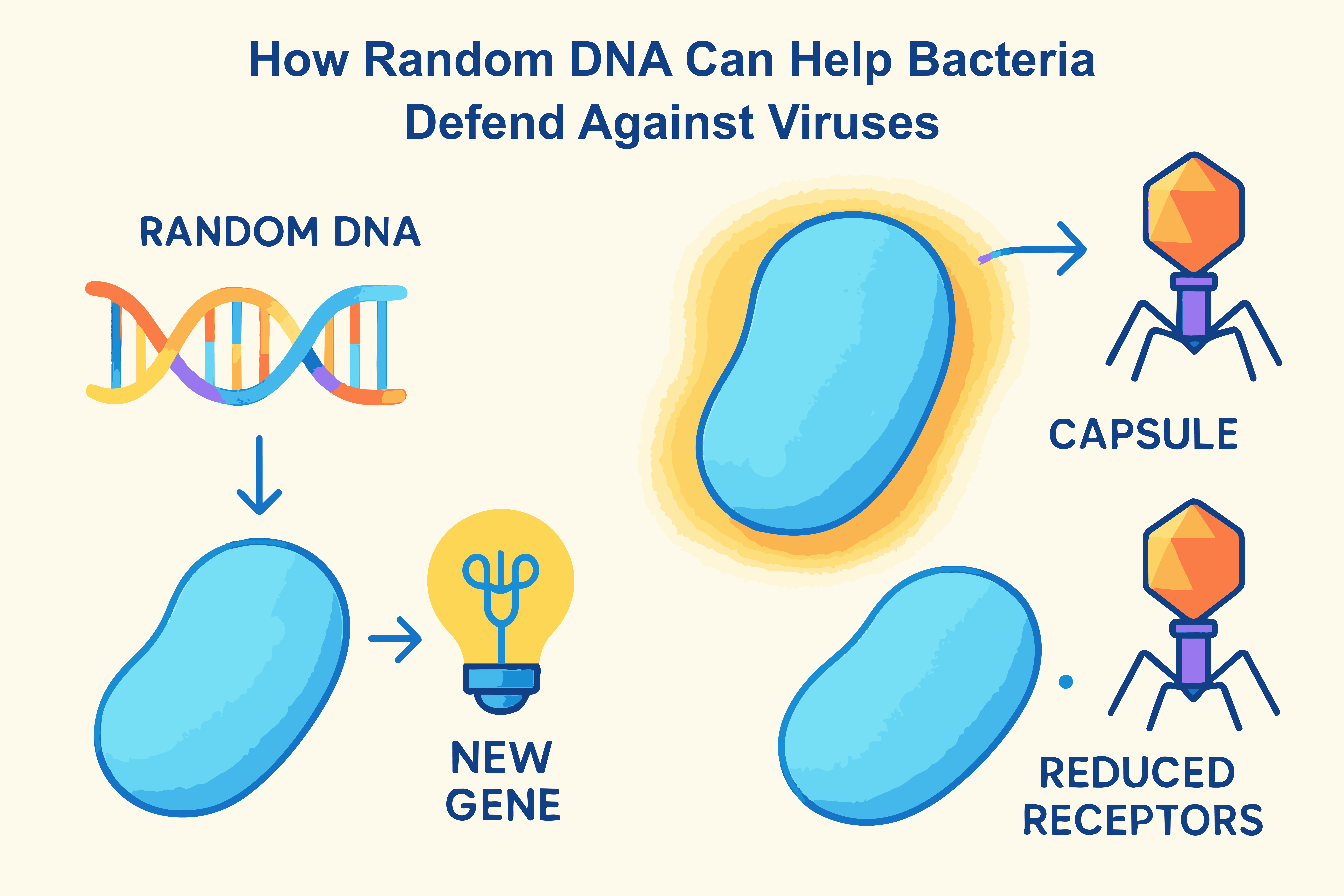
One of the most striking findings is that many random sequences protected E. coli from a diverse range of viruses by activating a natural stress-response pathway. This pathway triggers the production of a protective capsule on the cell surface, physically blocking viral infection. Even though these sequences were random, they could plug directly into an existing cellular network and “switch on” an antiviral defense system, demonstrating how easily evolution can stumble upon molecular solutions hidden in the vast space of possible sequences.
A second group of random sequences conferred highly specific resistance to the iconic T4 bacteriophage, one of the best-studied viruses in biology. These sequences did not create new antiviral proteins but instead subtly reduced the levels of a single bacterial surface receptor required for the virus to attach. This elegant mechanism shows that entirely new genes can influence vital cellular pathways and fine-tune gene expression, even without any prior evolutionary optimization. It also highlights a surprisingly simple route by which new antiviral traits may evolve in nature.
The study also reveals the dynamic nature of microbial evolution: when confronted with these newly emerged defenses, T4 phages rapidly evolved counter-adaptations. Within a single lab experiment, viruses acquired mutations that restored their ability to infect bacteria expressing the random resistance genes. This rapid arms race, innovation followed by counter-innovation, mirrors the evolutionary battles that have shaped microbial life for billions of years.
Overall, this work provides compelling evidence that functional novelty can emerge with surprising ease from random DNA, offering new insight into how genes originate and how organisms adapt to their ever-changing environments. It suggests that the birth of new genes may, in fact, be a powerful driver of evolution. These findings not only reshape our understanding of gene evolution but also open new possibilities for biotechnology, synthetic biology, and the engineering of antiviral defenses.


































