How perfect can interactomes be? Protein interactions underpin almost all of the processes that are required for life. One example is the chain of interactions between MAPK kinases that controls cell growth and proliferation, among many other functions. When imagining how such pathways may have evolved, one typically thinks of how the components evolved to ensure that protein 'A' can activate protein 'B', protein 'B' can activate protein 'C', etc., until a cell response is reliably achieved from an input hormone or signal.
It is often forgotten that proteins and their pathways do not evolve in isolation but inside cells that may be packed with thousands of other proteins. When proteins evolve, they grow not only to bind to their functional partners, but also to avoid functionally unrelated proteins. When non-cognate proteins interact without any functional benefit, it is called a 'spurious' interaction. An increasing amount of data over the past 15 years has highlighted the prevalence of such spurious interactions.
The HFSP Research Grant team, led by Christian Landry, Institut de biologie intégrative et des systèmes, Canada, is working on understanding how these unintended interactions affect the evolution of cell networks by first quantifying the impact of spurious interactions. For this, the research team turns towards the baker's yeast Saccharomyces cerevisiae – a model organism for synthetic biology. Quite simply, the scientists engineer a new physical interaction inside of yeast and then measure the effect of this on yeast growth (i.e., fitness). In the paper published on the EMBO Journal, HFSP researchers do this by expressing a family of kinases that never evolved in yeast, generating thousands of new (functionless) phosphosites not seen in the native yeast proteome.
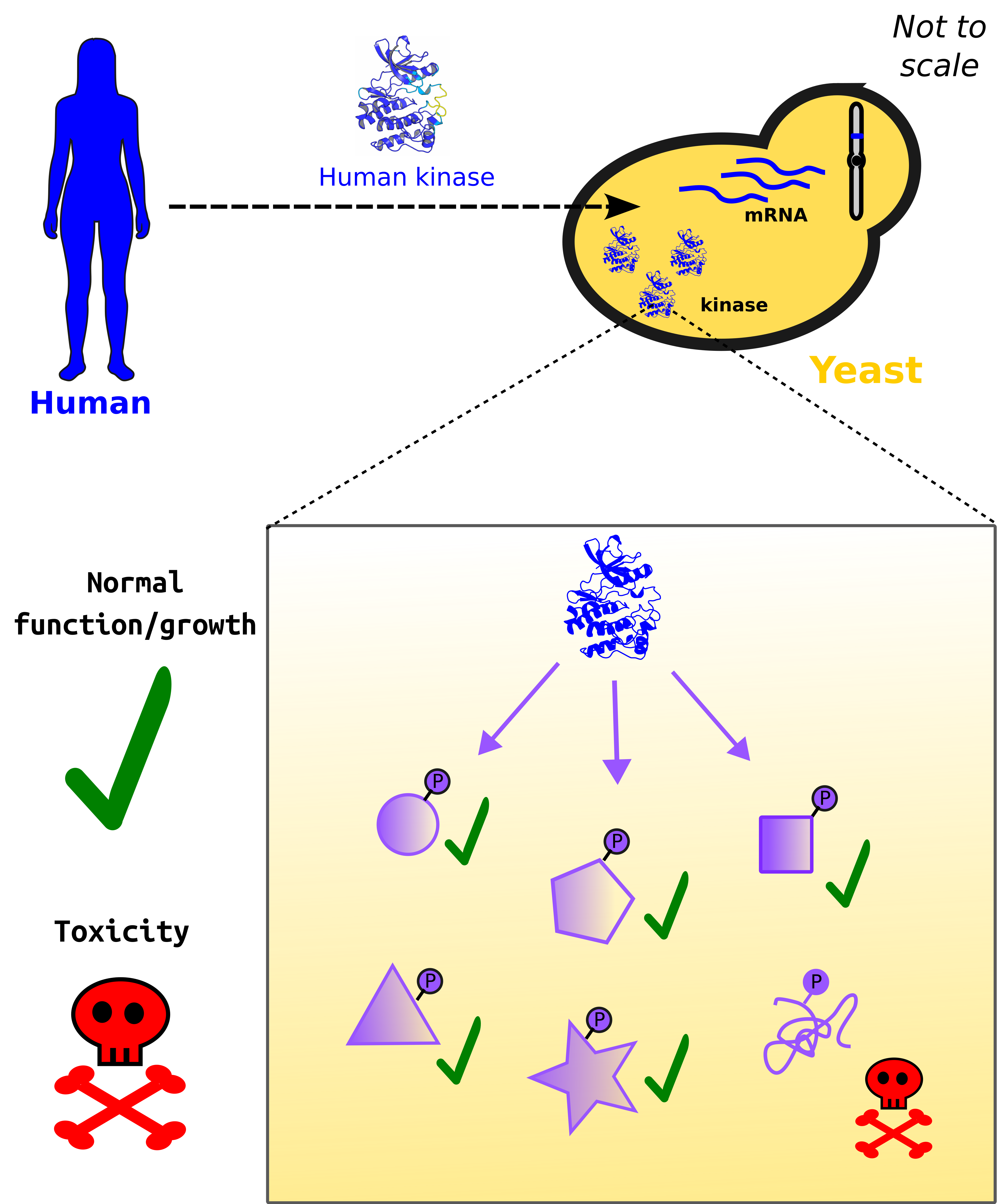
Landry's team found a strong negative correlation between the number of new phosphosites and fitness. That is, yeast strains tend to grow less well as more spurious phosphosites are added. However, one yeast strain can tolerate around 800 spurious phosphosites (a huge number compared to most perturbations) without any effect on fitness – this indicates that many of the 'new' interactions are harmless!
What is the precise relationship between spurious interactions and fitness? This is an open question for the scientists, but one possibility is that the cell can tolerate a sea of functionless 'noise' in the interactome space. Still, a small number of critical interactions may be highly toxic.
Much of the analysis performed by the HFSP Research Grant team was also computational. For example, they used AlphaFold to map the spurious phosphosites to around 2000 proteins belonging to baker's yeast. From this, researchers predict hundreds of protein structures to be destabilised by spurious phosphorylation. The effect on fitness ends up being less than expected. One possible reason is that for any given phosphorylated protein, there is still a large percentage of unmodified protein sufficient to perform the WT function.


































