An emerging paradigm regarding the assembly of protein molecules in cells is that the non-equilibrium nature of translation-related processes plays a key role in governing nascent protein behavior, including whether they co-translationally fold and function or misfold and malfunction. Several research papers have been published on this topic over the last decade. However, a number of fundamental questions remain unanswered, including what are the factors that determine why synonymous mutations at some codon positions in a transcript can have a large impact on a co-translational process, while at other positions there is no effect? Exactly how robust are co-translational processes to changes in codon translation rates? And, since codon usage helps to guide nascent protein behavior, can we learn the governing principles sufficiently well such that we can rationally design mRNA sequences to control nascent protein behavior?
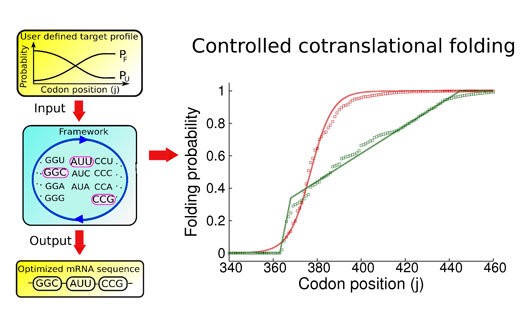
Figure: Illustration of the Monte Carlo master equation based framework with an example of the two user defined co-translational profiles of the TRXB protein. Solid lines are the user defined target profile for which we want our framework to optimize the mRNA sequence while discrete data points are the co-translational profiles obtained by using the optimized mRNA sequences produced by the framework.
We answer these questions by introducing a unique method for designing mRNA sequences to control co-translational protein folding in a user-defined manner. We verify this method against the results from molecular dynamics simulations of the translation process. Through careful analysis of these simulations, we demonstrate that codon positions that strongly influence nascent protein behavior are determined by how far a protein’s co-translational folding behavior is from equilibrium, and that the robustness of co-translational folding to single-point synonymous mutations is fundamentally related to the number of different synonymous mRNA sequences that can give rise to it.
The results also suggest experiments that scientists can carry out to identify putative critical codon positions and estimate the relative number of synonymous mRNA sequences that can give rise to a particular nascent protein behavior. Our chemical kinetic-based method is the first to explicitly connect the microscopic kinetic details of translation to nascent protein behavior, providing a novel tool to design mRNA sequences to finely control co-translational folding.
The O’Brien and Bukau Labs were awarded an HFSP grant in 2015 titled “Quantifying and predicting the influence of translation kinetics on nascent proteome behavior”. This paper achieves one of the aims of the proposal to understand the physical origins of the influence of translation kinetics on newly synthesized proteins.
Reference
Physical Origins of Codon Positions That Strongly Influence Cotranslational Folding: A Framework for Controlling Nascent-Protein Folding. Sharma AK, Bukau B, O’Brien EP (2016) J. Am. Chem. Soc., 138: 1180-1195.


































