Cells have evolved complex regulatory systems to adapt in fluctuating environments. At the heart of these circuits are proteins known as transcription factors, which behave as biosensors that control gene expression in response to changes in intracellular concentrations. In the new era of synthetic biology, biosensors are key components for reprogramming cells, but fine-tuning cell behavior is limited by our poor understanding of their tunability.
Biosensors bind to an inducing metabolite and alter gene expression by binding to the DNA. Their overall input-output behavior can be represented by a dose-response curve. Two key features of response curves are their dynamic range and sensing threshold. These two parameters determine the strength of biosensor output for various input concentrations. In synthetic biology, genetic engineering is typically employed to shape dose-response curves to a desired specification. Our experiments revealed that changes to the sensor-DNA interaction affect both dynamic range and threshold simultaneously. This indicates the existence of trade-offs between the ability to either sense low chemical abundance or increase biosensor output. We used mathematical models to elucidate the interdependencies between the dose-response curves and genetic modifications. Combining model analysis and genetic engineering, we found two tuning dials that allow for independent control of the dose-response parameters.
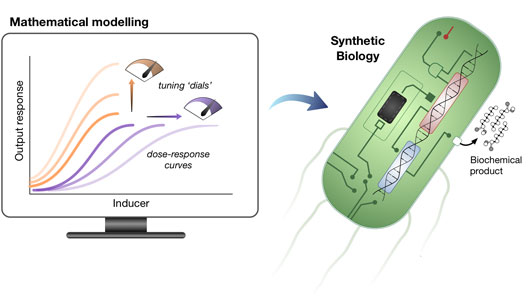
Figure: The synergy between mathematical models and synthetic biology facilitates the design of metabolite biosensors.
These findings are crucial for fine-tuning biosensor function through genetic and protein engineering, with applications in basic science as well as the construction of efficient microbial factors for the production of commodity chemicals. Our HFSP Young Investigator Grant was instrumental to start this exciting collaboration between mathematicians and synthetic biologists.
Reference
Fundamental Design Principles for Transcription-Factor-Based Metabolite Biosensors. Ahmad A. Mannan, Di Liu, Fuzhong Zhang, and Diego Oyarzún. ACS Synthetic Biology, Article ASAP, August 1, 2017 (web), DOI: 10.1021/acssynbio.7b00172.


































