The ability to distinguish self from non-self is an essential property of every immune system. Understanding the relationship between the efficiency with which immune systems recognize and destroy pathogens and, on the other hand, tolerate the host’s own biological components in order to prevent autoimmunity is fundamental to our understanding of how immune systems have evolved. However, experimental studies of this topic have remained elusive, mostly due to the complexity of immune systems in higher organisms. Remarkably, bacteria have evolved both innate and adaptive immune systems, and these lend themselves much more readily to experiments. We have been studying the simplest mechanism of bacterial innate immunity. Restriction-modification (RM) systems are minimally composed of two enzymes. The R enzyme cleaves DNA at specific target sites known as ‘restriction sites’. The M enzyme methylates the bacterium’s own restriction sites and thus prevents them from cleavage by R. When foreign pathogens, such as bacterial viruses or ‘bacteriophages’, infect the bacterial cell, their DNA typically contains several unmethylated restriction sites, which are rapidly cleaved by R and the infection is prevented. RM systems thus represent a minimal mechanism of self/non-self discrimination, performing this critical function with only two components.
 Because autoimmunity likely bears a significant cost in fitness, its frequency in populations is expected to be minimal. Although RM systems have been known and extensively studied for more than half a century, no negative fitness effects of a steadily maintained RM system have been experimentally detected. However, bioinformatic analyses of prokaryotic genomes detected a strong evolutionary signal in the form of a depletion of restriction sites, indicating that autoimmunity due to RM systems indeed occurs and is selected against, leading to restriction site avoidance. We recently employed highly sensitive methods to measure the frequency and effect of such autoimmunity events in bacteria at both population and single-cell level. We focused on two of the best-studied RM systems originating from Escherichia coli: EcoRI and EcoRV, and asked whether they induce autoimmunity at detectable rates. Our results show that about one third of one percent of all cells that carry the EcoRI RM system do experience autoimmunity, and their DNA is inadvertently cleaved. We detected no such effect for EcoRV.
Because autoimmunity likely bears a significant cost in fitness, its frequency in populations is expected to be minimal. Although RM systems have been known and extensively studied for more than half a century, no negative fitness effects of a steadily maintained RM system have been experimentally detected. However, bioinformatic analyses of prokaryotic genomes detected a strong evolutionary signal in the form of a depletion of restriction sites, indicating that autoimmunity due to RM systems indeed occurs and is selected against, leading to restriction site avoidance. We recently employed highly sensitive methods to measure the frequency and effect of such autoimmunity events in bacteria at both population and single-cell level. We focused on two of the best-studied RM systems originating from Escherichia coli: EcoRI and EcoRV, and asked whether they induce autoimmunity at detectable rates. Our results show that about one third of one percent of all cells that carry the EcoRI RM system do experience autoimmunity, and their DNA is inadvertently cleaved. We detected no such effect for EcoRV.
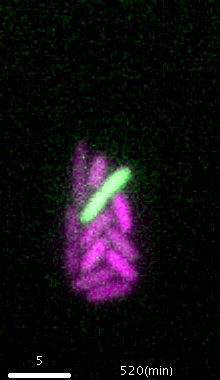
Figure: Escherichia coli cells growing inside a microfluidic device. The cell shown in green induces SOS response as a result of autoimmunity caused by the EcoRI RM system.
We found that the fitness cost of individual autoimmunity events is below the detection limit in bacteria proficient in DNA repair growing in resource-rich environments. However, the fitness cost increases significantly when resources are limited, or when DNA repair is impaired. These results suggested that the cost of autoimmunity is genetically and environmentally dependent. We further showed that EcoRV, which does not induce autoimmunity at a detectable rate, is less efficient at protecting the host from viral infections than EcoRI, pointing towards the existence of an evolutionary tradeoff between the efficiency of an immune system, and the risk of autoimmunity.
Since restriction sites of EcoRI, but not EcoRV, are under-represented in the genome of E. coli, our results supported the hypothesis that restriction site avoidance is an evolutionary consequence of bacterial autoimmunity due to RM systems. Our bioinformatics analysis revealed that some types of RM systems are on average more avoided than others. Specifically, RM systems, in which R and M are fused into a single peptide unit, are less avoided than the classical RM systems, in which R and M are structurally and functionally independent enzymes. Unlike in RM systems with fused components, in which the ratio of R and M enzymatic activities is constant, the ratio of enzymatic activities in structurally independent RM systems may be disturbed by stochastic events occurring at the level of single cells, thus increasing the frequency of autoimmunity. Our results identify rare stochastic events of autoimmunity caused by RM systems as a force shaping prokaryotic genomes, and improve our understanding of evolutionary tradeoffs in bacterial immune systems.
Reference
Bacterial Autoimmunity Due to a Restriction-Modification System. Pleška M, Qian L, Okura R, Bergmiller T, Wakamoto Y, Kussell E, and Guet CC. Current Biology 26, 404–409 (2016).
Nature Reviews Microbiology Research Highlight


































