Cells destroy unwanted proteins to control protein levels and remove those that are damaged. Using an enzyme complex called the proteasome, these unwanted proteins are broken down, eventually releasing amino acids that can be recycled to make new proteins. Deficient proteasome function leads to the build-up of unneeded, damaged, and potentially toxic proteins. To prevent this, cells can compensate for a shortage in proteasome function by producing more proteasomes. In C. elegans, this increase in proteasome levels was known to result from the activation of a transcription factor called SKN-1, which controls expression of proteasome subunit genes. How SKN-1 senses a reduction in proteasome function to trigger this response was not known.
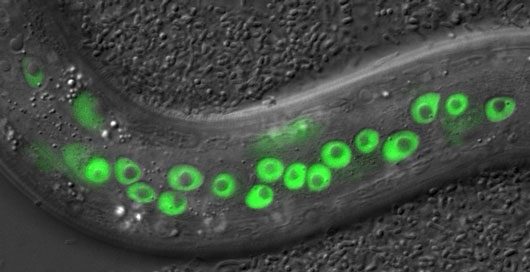
Figure: Image showing expression of the DDI-1 protease (shown in green) in C. elegans during proteasome dysfunction. In these animals DDI-1 localizes to nuclei where it cleaves and activates SKN-1.
HFSP Long-Term Fellow Nicolas Lehrbach carried out a large-scale genetic screen to identify C. elegans mutants that fail to activate SKN-1 in response to proteasome dysfunction. Among the many mutations found in this screen were several affecting a protease enzyme called DDI-1, which is found in many species, including humans, but had not previously been linked to regulation of proteasome levels. Another factor identified by this screen was PNG-1, an enzyme that removes attached sugar molecules from proteins; a process called deglycosylation. Lehrbach showed that in order for SKN-1 to become active, it must be deglycosylated by PNG-1, and cleaved by the DDI-1 protease. As a result, mutant animals that lack PNG-1 or DDI-1 cannot compensate for diminished proteasome function and are highly sensitive to environmental or genetic insults to the proteasome.
In another paper published simultaneously in eLife, a team lead by Shigeo Murata at the University of Tokyo independently discovered that the human version of DDI-1, called DDI2, is needed for human cells to increase proteasome levels in response to proteasome dysfunction. Together, these two papers suggest that the DDI-1/2 protease is an essential regulator of the proteasome in animals from worms to humans.
This work suggests new avenues for treatment of diseases in which proteasome activity is either too high or too low. Boosting proteasome activity might help to treat neurodegenerative diseases such as Alzheimer’s and Parkinson’s, in which accumulation of unwanted proteins disrupts neuronal function. Proteasome inhibitor drugs are used to treat cancers such as multiple myeloma, but the cancer cells’ ability to compensate by increasing proteasome levels limits the effectiveness of this treatment. Drugs that target DDI-1 might overcome this problem if administered in combination with proteasome inhibitors. Deleterious mutations in NGLY1, the human version of PNG-1, have recently been found to underlie a rare genetic disease. The finding that PNG-1 is needed to regulate proteasome levels provides an explanation for the symptoms caused by NGLY1 deficiency. It will now be possible, using model systems such as C. elegans, to search for ways to restore normal regulation of proteasomes in NGLY1 patients.
Reference
Proteasome dysfunction triggers activation of SKN-1A/Nrf1 by the aspartic protease DDI-1. Lehrbach NJ and Ruvkun G. eLife. 2016;5:e17721
Other references
The aspartyl protease DDI2 activates Nrf1 to compensate for proteasome dysfunction. Shun Koizumi, Taro Irie, Shoshiro Hirayama, Yasuyuki Sakurai, Hideki Yashiroda, Isao Naguro, Hidenori Ichijo, Jun Hamazaki, Shigeo Murata. eLife. 2016;5:e18357.


































