Nearly one quarter of the genes in all organisms encode membrane proteins. This vast group of proteins performs an abundance of functions and is involved in just about every physiological process and their malfunction is the basis for a plethora of diseases. In spite of their importance, methods for studying membrane proteins have lagged far behind their soluble protein counterparts, leaving many aspects of membrane protein biology incompletely understood. One central question that remains open is how membrane proteins fold in vivo.
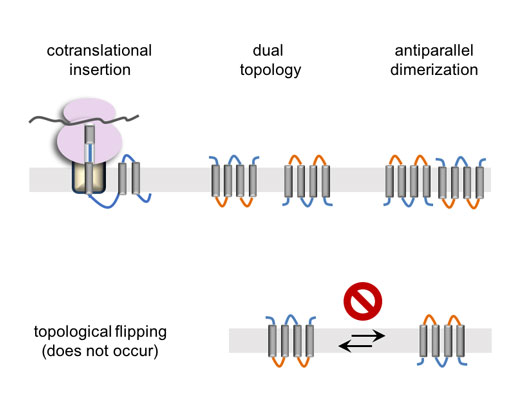
Figure: Membrane protein biogenesis and (dual) topology generation.
The largest class of membrane proteins is comprised of transmembrane α-helix bundles, in which the polypeptide crosses the membrane several times using hydrophobic α-helices. The transmembrane α-helices are connected by relatively hydrophilic loops that stick out of both sides of the membrane (see figure). These structures need to be inserted into the membrane in the right orientation, posing a challenge for the cell. Consequently, the biogenesis of membrane proteins is quite different from that of water-soluble ones. Being very hydrophobic, they are extremely prone to aggregation and therefore must be inserted into the membrane co-translationally, as the transmembrane helices emerge from the ribosome (see figure). At the end of this co-translational insertion process, the membrane topology is already acquired, namely which parts of the polypeptide are in or out of the membrane and what orientation the protein takes in the membrane.
Once attained, the topologies of membrane proteins have generally been assumed to be fixed. This is because any change in the topology would require transferring the hydrophilic loops through the hydrophobic membrane barrier. This view, however, has recently been challenged by results from several model proteins, which point to a more dynamic behavior, in which post-translational changes in the topology are possible. This kind of dynamics may open a plethora of unexpected possibilities. For example, it may be possible that not all helices have to be inserted co-translationally, which may help solving a long-standing conundrum of how more hydrophilic transmembrane helices get inserted. Membrane proteins could perhaps also change their topologies dynamically in response to different physiological states to regulate their function. Therefore, understanding to what extent such dynamics are possible is central to our understanding of membrane protein biology.
In a recently published study, we examined the potential for membrane proteins to flip their membrane orientation in vivo. As a model, we took advantage of dimeric dual topology proteins, a model system that is unique in several ways. First, these are relatively small proteins, with only 4 helices and quite short loops, making reorientation theoretically quicker than for almost all other membrane proteins. Secondly, the proteins have dual topology, a curious rarity among membrane proteins, meaning that they exist in equal numbers in both orientations, which dimerize together to form anti-parallel homodimers. This means that evolution may have selected these proteins to be dynamic, such that they may not have to be inserted in a perfectly balanced ratio of the two different topologies from the outset. According to this scenario, it may not matter in which orientation the protein inserts, since it would simply flip-flop back and forth until all subunits dimerize with oppositely-oriented partners, thereby fixing the final dual topology. It therefore seems likely that if membrane proteins could flip their orientations in the membrane, so should dual topology dimers.
In the study, we tested whether dimerization has any effect on orientation, by mapping the orientations of different mutants in which the orientation or the dimerization are disrupted. But no matter how the system was manipulated, dimerization had no effect on topology. The most likely explanation is that by the time protein subunits are available for dimerization, it is too late for them to change orientations. The results therefore suggest that membrane proteins are probably ‘stuck’ in the initial orientation they were inserted in, suggesting that, as thought before, the membrane poses a strong kinetic constraint on membrane protein dynamics.
The author would like to thank Gunnar von Heijne and Grant Kemp for their assistance in preparing this article.
Reference
Stable membrane orientations of small dual-topology membrane proteins. Fluman N, Tobiasson V and von Heijne G. Proc Natl Acad Sci U S A. 2017 Jul 25;114(30):7987-7992.


































